
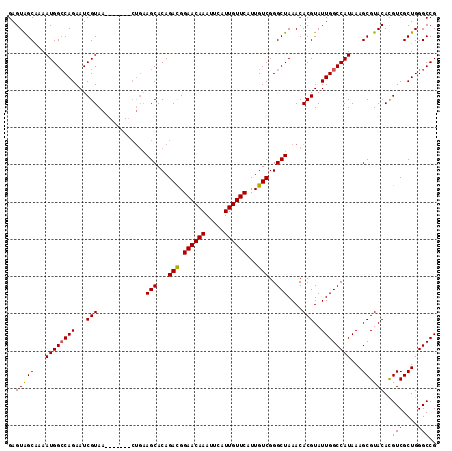
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,906,922 – 24,907,088 |
| Length | 166 |
| Max. P | 0.991798 |

| Location | 24,906,922 – 24,907,030 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.70 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -26.77 |
| Energy contribution | -26.65 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24906922 108 + 27905053 GAGUAGCAAAAUGGGCAGAAUCGUAA-------CUGAAGCACAGACGGAACAAAUUCAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACAUGUCGCUGGGCCG (..((((..(((((((((...(((..-------(((.....))))))(((....))).))))))))).....))))..).......((((....((((.......)))).)))). ( -28.00) >DroSec_CAF1 18596 108 + 1 GAGUAGCAAAAUGGCCAGAAUCGUAA-------CUGAAGCACAGACGGAACAAAUUCAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCG (..((((...((((((((...(((..-------....(((...(((.((((((.....))))))...)))..)))....)))..))))))))...(((....))).))))..).. ( -31.20) >DroSim_CAF1 18235 108 + 1 GAGUGGCAAAAUGGCCAGAAUCGUAA-------CUGAAGCACAGACGGAACAAAUUCAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCG ....(((...((((((((...(((..-------....(((...(((.((((((.....))))))...)))..)))....)))..))))))))..((((.......))))..))). ( -34.90) >DroEre_CAF1 18684 110 + 1 GAGUAGCGAAAUGGCCAGAAUCGUAACUCGUA-----AGCACAGAUGGAACAAAUUUAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCG ((((.((((...........)))).))))...-----(((...(((.((((((.....))))))...)))..)))...........((((....((((.......)))).)))). ( -31.70) >DroYak_CAF1 18921 114 + 1 GAGUAGCGAAAUGGCCAGAAUCGUAA-UCGUAACUGAAGCACAGACGGAACAAAUUUAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCG (..((((((.((((((((...(((..-(((....)))(((...(((.((((((.....))))))...)))..)))....)))..))))))))....((....))))))))..).. ( -33.30) >consensus GAGUAGCAAAAUGGCCAGAAUCGUAA_______CUGAAGCACAGACGGAACAAAUUCAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCG ..(((((...((((((((...(((.............(((...(((.((((((.....))))))...)))..)))....)))..))))))))...)).)))..(((....))).. (-26.77 = -26.65 + -0.12)


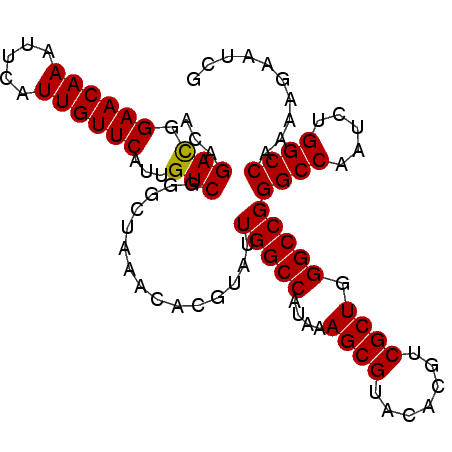
| Location | 24,906,955 – 24,907,053 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 97.76 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -29.46 |
| Energy contribution | -29.30 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24906955 98 + 27905053 ACAGACGGAACAAAUUCAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACAUGUCGCUGGGCCGGGCCAAUCUGGCCAAAAGAAUCG ...(((.((((((.....))))))...))).((((((.......))))))....((((.......)))).(((((((....))))))).......... ( -30.10) >DroSec_CAF1 18629 98 + 1 ACAGACGGAACAAAUUCAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCGGGCCAAAAUGGCCAAAAGAAUCG ...(((.((((((.....))))))...)))...............(((((....((((.......)))).)))))((((.....)))).......... ( -29.60) >DroSim_CAF1 18268 98 + 1 ACAGACGGAACAAAUUCAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCGGGCCAAUCUGGCCAAAAGAAUCG ...(((.((((((.....))))))...))).((((((.......))))))....((((.......)))).(((((((....))))))).......... ( -30.10) >DroEre_CAF1 18719 98 + 1 ACAGAUGGAACAAAUUUAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCGGGCCAAUCUGGCCAAAAGAAUCG ...(((.((((((.....))))))...))).((((((.......))))))....((((.......)))).(((((((....))))))).......... ( -28.60) >DroYak_CAF1 18960 98 + 1 ACAGACGGAACAAAUUUAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCGGGCCAAUCUGGCCAAAAGAAUCG ...(((.((((((.....))))))...))).((((((.......))))))....((((.......)))).(((((((....))))))).......... ( -30.10) >consensus ACAGACGGAACAAAUUCAUUGUUCAUUGUCGGGCUAAACACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCGGGCCAAUCUGGCCAAAAGAAUCG ...(((.((((((.....))))))...)))...............(((((....((((.......)))).)))))((((.....)))).......... (-29.46 = -29.30 + -0.16)


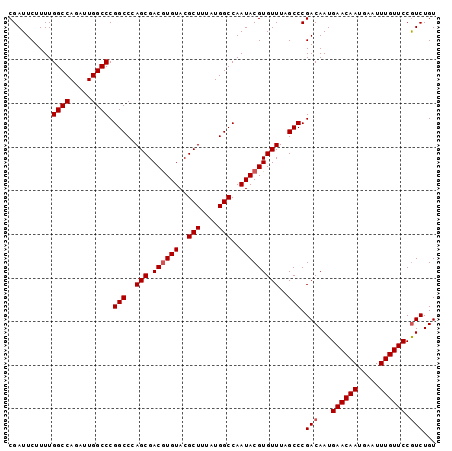
| Location | 24,906,955 – 24,907,053 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 97.76 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -26.86 |
| Energy contribution | -27.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24906955 98 - 27905053 CGAUUCUUUUGGCCAGAUUGGCCCGGCCCAGCGACAUGUACGCUUUAUGGCCAAUACGUGUUUAGCCCGACAAUGAACAAUGAAUUUGUUCCGUCUGU ..........((((.....)))).(((.....((((((((.(((....)))...))))))))..))).(((...((((((.....)))))).)))... ( -27.00) >DroSec_CAF1 18629 98 - 1 CGAUUCUUUUGGCCAUUUUGGCCCGGCCCAGCGACGUGUACGCUUUAUGGCCAAUACGUGUUUAGCCCGACAAUGAACAAUGAAUUUGUUCCGUCUGU ..........((((.....)))).(((..(((.((((((..(((....)))..)))))))))..))).(((...((((((.....)))))).)))... ( -28.80) >DroSim_CAF1 18268 98 - 1 CGAUUCUUUUGGCCAGAUUGGCCCGGCCCAGCGACGUGUACGCUUUAUGGCCAAUACGUGUUUAGCCCGACAAUGAACAAUGAAUUUGUUCCGUCUGU ..........((((.....)))).(((..(((.((((((..(((....)))..)))))))))..))).(((...((((((.....)))))).)))... ( -28.80) >DroEre_CAF1 18719 98 - 1 CGAUUCUUUUGGCCAGAUUGGCCCGGCCCAGCGACGUGUACGCUUUAUGGCCAAUACGUGUUUAGCCCGACAAUGAACAAUAAAUUUGUUCCAUCUGU ((........((((.....)))).(((..(((.((((((..(((....)))..)))))))))..))))).....((((((.....))))))....... ( -26.50) >DroYak_CAF1 18960 98 - 1 CGAUUCUUUUGGCCAGAUUGGCCCGGCCCAGCGACGUGUACGCUUUAUGGCCAAUACGUGUUUAGCCCGACAAUGAACAAUAAAUUUGUUCCGUCUGU ..........((((.....)))).(((..(((.((((((..(((....)))..)))))))))..))).(((...((((((.....)))))).)))... ( -28.80) >consensus CGAUUCUUUUGGCCAGAUUGGCCCGGCCCAGCGACGUGUACGCUUUAUGGCCAAUACGUGUUUAGCCCGACAAUGAACAAUGAAUUUGUUCCGUCUGU ..........((((.....)))).(((..(((.((((((..(((....)))..)))))))))..))).(((...((((((.....)))))).)))... (-26.86 = -27.26 + 0.40)


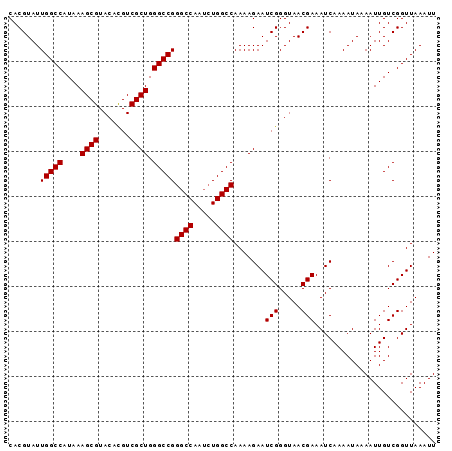
| Location | 24,906,993 – 24,907,088 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 98.74 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24906993 95 + 27905053 CACGUAUUGGCCAUAAAGCGUACAUGUCGCUGGGCCGGGCCAAUCUGGCCAAAAGAAUCGGGUAACGAAAUCAAAAUAAAAUUGUCGGUUAAAUU .......(((((....((((.......)))).)))))((((.....)))).......(((.....)))........................... ( -23.70) >DroSec_CAF1 18667 95 + 1 CACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCGGGCCAAAAUGGCCAAAAGAAUCGGGUAACGAAAUCAAAAUAAAAUUGUCGGUUAAAUU .......(((((....((((.......)))).)))))((((.....)))).......(((.....)))........................... ( -23.70) >DroSim_CAF1 18306 95 + 1 CACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCGGGCCAAUCUGGCCAAAAGAAUCGGGUAACGAAAUCAAAAUAAAAUUGUCGGUUAAAUU .......(((((....((((.......)))).)))))((((.....)))).......(((.....)))........................... ( -23.70) >DroEre_CAF1 18757 95 + 1 CACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCGGGCCAAUCUGGCCAAAAGAAUCGGGUAACGAAAUCAAAAUAAAAUUGUCGGUUAAAUU .......(((((....((((.......)))).)))))((((.....)))).......(((.....)))........................... ( -23.70) >DroYak_CAF1 18998 95 + 1 CACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCGGGCCAAUCUGGCCAAAAGAAUCGGGUAACGAAAUCAAAAUAAAAUUGUCGGUUAAAUU .......(((((....((((.......)))).)))))((((.....)))).......(((.....)))........................... ( -23.70) >consensus CACGUAUUGGCCAUAAAGCGUACACGUCGCUGGGCCGGGCCAAUCUGGCCAAAAGAAUCGGGUAACGAAAUCAAAAUAAAAUUGUCGGUUAAAUU .......(((((....((((.......)))).)))))((((.....)))).......(((.....)))........................... (-23.70 = -23.70 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:33:16 2006