
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,903,382 – 24,903,542 |
| Length | 160 |
| Max. P | 0.939410 |

| Location | 24,903,382 – 24,903,502 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -29.90 |
| Energy contribution | -30.06 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24903382 120 - 27905053 GCGGACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUGUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUGUAAGAAUUGAUGGCAAACAAUGCGCGAAUGGGCGGAGCAA ((....((((((((((....)))))))))).(((.(((..((((((...(((.(((((....))))).))).))))))..)))...)))...))......(.(((......))).).... ( -33.20) >DroSec_CAF1 16418 120 - 1 GCGUACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUGUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUGUAAGAAUUGAUGGCAAACAAUGCGCGAAUGGGCGGAGCAA (((((.((((((((((....)))))))))).(((.(((..((((((...(((.(((((....))))).))).))))))..)))...)))...........)))))............... ( -34.10) >DroSim_CAF1 15752 120 - 1 GCGUACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUGUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUGUAAGAAUUGAUGGCAAACAAUGCGCGAAUGGGCGGAGCAA (((((.((((((((((....)))))))))).(((.(((..((((((...(((.(((((....))))).))).))))))..)))...)))...........)))))............... ( -34.10) >DroEre_CAF1 16487 120 - 1 GCGGACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUCUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUGUAAGAAUUGAUGGCAAACAAUGCGCGAAUGGGCAGAGCAA ((....((((((((((....))))))))))......((((((((((...(((.(((((....))))).))).)))))(((((....(((....)))....)))))......))))))).. ( -33.40) >DroYak_CAF1 16631 120 - 1 UCGAACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUGUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUGUAAGAAUUGAUGGCAAACAAUGCGCGAAUGGGCAGUGCAA ((((..((((((((((....)))))))))).....(((..((((((...(((.(((((....))))).))).))))))..)))...))))..(((.....(((.(....).))).))).. ( -34.30) >consensus GCGGACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUGUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUGUAAGAAUUGAUGGCAAACAAUGCGCGAAUGGGCGGAGCAA ((....((((((((((....)))))))))).(((.(((..((((((...(((.(((((....))))).))).))))))..)))...)))...))......(((.(....).)))...... (-29.90 = -30.06 + 0.16)

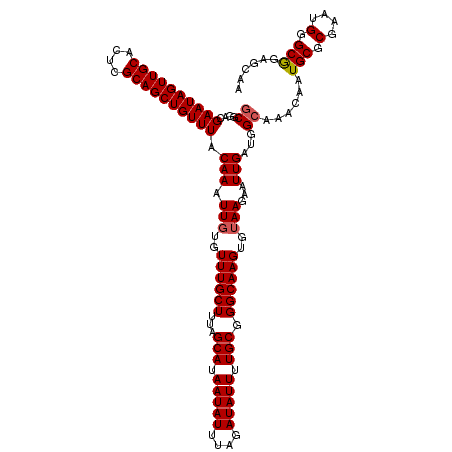
| Location | 24,903,422 – 24,903,542 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -30.94 |
| Energy contribution | -31.66 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24903422 120 - 27905053 CAAGUGGCAAGAAGCAGCGGUAGGAGGCCAAGUGCGAGGGGCGGACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUGUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUG ......((((.((((((((((.....)))...((((((..(((.((.....))))).))))))))))))).....)))).((((((...(((.(((((....))))).))).)))))).. ( -38.60) >DroSec_CAF1 16458 120 - 1 CAAGUGGCAAGAAGCAGCGGCAGGAGGCGGAAUGCGAGGGGCGUACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUGUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUG ......((((.(((((((.((.....))....((((((..(((.((.....))))).))))))))))))).....)))).((((((...(((.(((((....))))).))).)))))).. ( -36.80) >DroSim_CAF1 15792 120 - 1 CAAGUGGCAAGAAGCAGCGGCAGGAGGCGGAGUGCGAGGGGCGUACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUGUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUG ((..((.(..(((((((..((((.((((((..((((((..(((.((.....))))).)))))).)))))).....))))..))))))).(((.(((((....))))).))).).))..)) ( -37.40) >DroEre_CAF1 16527 120 - 1 CAAGUGGCAAGAAGCAGCGGAAGGAGGCGGAGUGCGAGGGGCGGACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUCUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUG ((..((.(..((((((((((((.(((((((..((((((..(((.((.....))))).)))))).)))))).)...))))).))))))).(((.(((((....))))).))).).))..)) ( -40.70) >DroYak_CAF1 16671 120 - 1 CAAGUGGCAAGAAGCAGCGGAAGGAGGCGGAGUGCGAGGGUCGAACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUGUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUG ......((((.(((((((........(((.(((((((...((....))....)))))))))).))))))).....)))).((((((...(((.(((((....))))).))).)))))).. ( -34.40) >consensus CAAGUGGCAAGAAGCAGCGGAAGGAGGCGGAGUGCGAGGGGCGGACGAAUAGUUGCACUCGCAGCUGUUUACAAAUUGUGUUUGCUUUAGCAUAAUAUUUAGAUAUUUUGCGGGCAAGUG ......((((.(((((((.((.....))....((((((..(((.((.....))))).))))))))))))).....)))).((((((...(((.(((((....))))).))).)))))).. (-30.94 = -31.66 + 0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:33:12 2006