
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,903,085 – 24,903,284 |
| Length | 199 |
| Max. P | 0.857886 |

| Location | 24,903,085 – 24,903,204 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -25.74 |
| Energy contribution | -26.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24903085 119 + 27905053 UUCACAAA-AUCUUUUCAGCAGAUCCUGCACUCCUUUCUAGUCUGUCGGCCGCCACUCGCACCACCGUCGAUAAUCGGAUUAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGAUGGCAA ........-.........((((...))))........(((..(((((((..((((.(((.((....))))).....(((((((.....)))))))....))))..)))))))..)))... ( -32.30) >DroSec_CAF1 16132 119 + 1 UUCACAAA-AUCUUUUCAGCAGAUCCUGCACUCCUUUCUUGUCUGUCGGCCGCCACUCGAACCACCGUCGAUAAUCGGAUUAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGACGGCAA ........-.........((((...)))).........(((((((((((..((((.((((.......)))).....(((((((.....)))))))....))))..)))))).)))))... ( -32.10) >DroSim_CAF1 15458 119 + 1 UUCACAAA-AUCUUUUCAGCAGAUCCUGCACUCCUUUCUUGUCUGUCGGCCGCCACUCGAACCACCGUCGAUAAUCGGAUUAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGACGGCAA ........-.........((((...)))).........(((((((((((..((((.((((.......)))).....(((((((.....)))))))....))))..)))))).)))))... ( -32.10) >DroEre_CAF1 16191 117 + 1 UUCACAAAAAUCUUUUCACUAGAUCCUGCACUCCUUUCUUGUCUGUCGGC---CACUCGAACCCCCGUCGAUAAGCGGAUUAGAAUUUCUGGUCCUAAUUGGCACUCGACAGGGCGGUAA .........((((.......)))).((((....(......)(((((((((---((.((((.......)))).....(((((((.....)))))))....))))...)))))))))))... ( -27.70) >DroYak_CAF1 16342 117 + 1 UUCACAAAUAUCUUUUCAGUAGAUCCUGCACUCCUUUCCUGUCUGUCGGC---CACUCGAACCCCCGUCGAUAAUCGGAUUAGAAUUUCUGGUCCUAAUUGGCACUCGACAGGGCGGUAA ........((((......((((...)))).......(((((((.(...((---((.((((.......)))).....(((((((.....)))))))....))))...)))))))).)))). ( -30.60) >consensus UUCACAAA_AUCUUUUCAGCAGAUCCUGCACUCCUUUCUUGUCUGUCGGCCGCCACUCGAACCACCGUCGAUAAUCGGAUUAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGACGGCAA ..................((((...)))).........(((((((((((..((((.(((.((....))))).....(((((((.....)))))))....))))..)))))).)))))... (-25.74 = -26.14 + 0.40)



| Location | 24,903,124 – 24,903,244 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.84 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -27.81 |
| Energy contribution | -29.01 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24903124 120 + 27905053 GUCUGUCGGCCGCCACUCGCACCACCGUCGAUAAUCGGAUUAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGAUGGCAAAACGGUUGCUCUCAAACCGCCACAAUUGCAGCCCCCUGUC .((((((((..((((.(((.((....))))).....(((((((.....)))))))....))))..))))))))..((((....((((((..................))))))...)))) ( -36.27) >DroSec_CAF1 16171 118 + 1 GUCUGUCGGCCGCCACUCGAACCACCGUCGAUAAUCGGAUUAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGACGGCAAAGCGGUUGCUC--CAACCGCCGCAAUUGCAGCCCCCUGUC (((((((((..((((.((((.......)))).....(((((((.....)))))))....))))..)))))).)))(((...(((((((...--))))))).((....)).)))....... ( -44.30) >DroSim_CAF1 15497 118 + 1 GUCUGUCGGCCGCCACUCGAACCACCGUCGAUAAUCGGAUUAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGACGGCAAAGCGGUUGCUC--CAACCGCCGCAAUUGCAGCCCCCUGUC (((((((((..((((.((((.......)))).....(((((((.....)))))))....))))..)))))).)))(((...(((((((...--))))))).((....)).)))....... ( -44.30) >DroEre_CAF1 16231 115 + 1 GUCUGUCGGC---CACUCGAACCCCCGUCGAUAAGCGGAUUAGAAUUUCUGGUCCUAAUUGGCACUCGACAGGGCGGUAAACCGUUUGCUC--AAACCGCCACAAUUGCAGCCCCCUGUC ..((((((((---((.((((.......)))).....(((((((.....)))))))....))))...))))))((((((.............--..))))))......((((....)))). ( -35.76) >DroYak_CAF1 16382 97 + 1 GUCUGUCGGC---CACUCGAACCCCCGUCGAUAAUCGGAUUAGAAUUUCUGGUCCUAAUUGGCACUCGACAGGGCGGUAAACCGUUU--------------------GCAGCCCCCUGUC ((((((((((---.............)))))))...(((((((.....))))))).....)))....(((((((.(((.........--------------------...)))))))))) ( -31.52) >consensus GUCUGUCGGCCGCCACUCGAACCACCGUCGAUAAUCGGAUUAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGACGGCAAACCGGUUGCUC__AAACCGCCACAAUUGCAGCCCCCUGUC ..(((((((..((((.(((.((....))))).....(((((((.....)))))))....))))..)))))))(((((......((((((..................))))))..))))) (-27.81 = -29.01 + 1.20)



| Location | 24,903,164 – 24,903,284 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.06 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -26.31 |
| Energy contribution | -27.11 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24903164 120 + 27905053 UAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGAUGGCAAAACGGUUGCUCUCAAACCGCCACAAUUGCAGCCCCCUGUCUUUUUGAUGUUUGGCCACAAAUUCUGGACAAUGGCCAAGC ..........(((((.....)).))).((((((..(((.....((((.......))))((.......)).))).))))))........(((((((((..............))))))))) ( -33.44) >DroSec_CAF1 16211 118 + 1 UAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGACGGCAAAGCGGUUGCUC--CAACCGCCGCAAUUGCAGCCCCCUGUCUUUUUGAUGUUUGGCCACAAAUUCUGGACAAUGGCCAAGC ..........(((((.....)).))).((((((..(((...(((((((...--))))))).((....)).))).))))))........(((((((((..............))))))))) ( -40.44) >DroSim_CAF1 15537 118 + 1 UAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGACGGCAAAGCGGUUGCUC--CAACCGCCGCAAUUGCAGCCCCCUGUCUUUUUGAUGUUUGGCCACAAAUUCUGGACAAUGGCCAAGC ..........(((((.....)).))).((((((..(((...(((((((...--))))))).((....)).))).))))))........(((((((((..............))))))))) ( -40.44) >DroEre_CAF1 16268 118 + 1 UAGAAUUUCUGGUCCUAAUUGGCACUCGACAGGGCGGUAAACCGUUUGCUC--AAACCGCCACAAUUGCAGCCCCCUGUCUUUUUCAUGUUUGGCCACAAAUUCUGGACAAUGGCCAAGC ..(((.....(((((.....)).))).(((((((.(((.....((((....--)))).((.......)).))))))))))...)))..(((((((((..............))))))))) ( -33.84) >DroYak_CAF1 16419 100 + 1 UAGAAUUUCUGGUCCUAAUUGGCACUCGACAGGGCGGUAAACCGUUU--------------------GCAGCCCCCUGUCUUUUUCAUGUUUGGCCACAAAUUCUGGACAAUGGCCAAGC ..(((.....(((((.....)).))).(((((((.(((.........--------------------...))))))))))...)))..(((((((((..............))))))))) ( -30.74) >consensus UAGAUUUUCUGGUCCUAAUUGGCACUCGACAGGACGGCAAACCGGUUGCUC__AAACCGCCACAAUUGCAGCCCCCUGUCUUUUUGAUGUUUGGCCACAAAUUCUGGACAAUGGCCAAGC ..........(((((.....)).))).((((((..(......)((((((..................)))))).))))))........(((((((((..............))))))))) (-26.31 = -27.11 + 0.80)

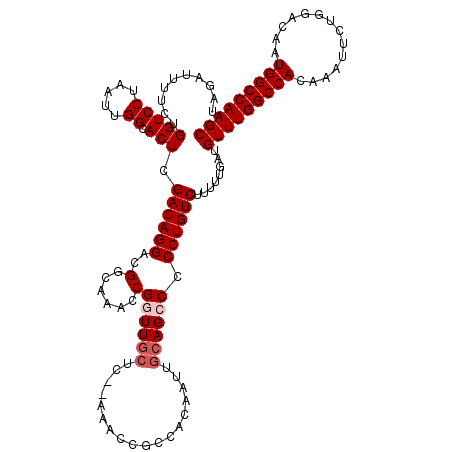

| Location | 24,903,164 – 24,903,284 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.06 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -29.28 |
| Energy contribution | -31.28 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24903164 120 - 27905053 GCUUGGCCAUUGUCCAGAAUUUGUGGCCAAACAUCAAAAAGACAGGGGGCUGCAAUUGUGGCGGUUUGAGAGCAACCGUUUUGCCAUCCUGUCGAGUGCCAAUUAGGACCAGAAAAUCUA (.((((((((............)))))))).).((.....((((((((((.((....))((((((((....).)))))))..))).)))))))(.((.((.....))))).))....... ( -36.00) >DroSec_CAF1 16211 118 - 1 GCUUGGCCAUUGUCCAGAAUUUGUGGCCAAACAUCAAAAAGACAGGGGGCUGCAAUUGCGGCGGUUG--GAGCAACCGCUUUGCCGUCCUGUCGAGUGCCAAUUAGGACCAGAAAAUCUA (.((((((((............)))))))).).((.....((((((((((.((....))((((((((--...))))))))..))).)))))))(.((.((.....))))).))....... ( -42.00) >DroSim_CAF1 15537 118 - 1 GCUUGGCCAUUGUCCAGAAUUUGUGGCCAAACAUCAAAAAGACAGGGGGCUGCAAUUGCGGCGGUUG--GAGCAACCGCUUUGCCGUCCUGUCGAGUGCCAAUUAGGACCAGAAAAUCUA (.((((((((............)))))))).).((.....((((((((((.((....))((((((((--...))))))))..))).)))))))(.((.((.....))))).))....... ( -42.00) >DroEre_CAF1 16268 118 - 1 GCUUGGCCAUUGUCCAGAAUUUGUGGCCAAACAUGAAAAAGACAGGGGGCUGCAAUUGUGGCGGUUU--GAGCAAACGGUUUACCGCCCUGUCGAGUGCCAAUUAGGACCAGAAAUUCUA ...............(((((((.(((((............(((((((.((..(....)..))(((..--((((.....))))))).)))))))............)).))).))))))). ( -37.05) >DroYak_CAF1 16419 100 - 1 GCUUGGCCAUUGUCCAGAAUUUGUGGCCAAACAUGAAAAAGACAGGGGGCUGC--------------------AAACGGUUUACCGCCCUGUCGAGUGCCAAUUAGGACCAGAAAUUCUA ...............(((((((.(((((............(((((((((..((--------------------.....))...)).)))))))............)).))).))))))). ( -29.35) >consensus GCUUGGCCAUUGUCCAGAAUUUGUGGCCAAACAUCAAAAAGACAGGGGGCUGCAAUUGCGGCGGUUG__GAGCAACCGCUUUGCCGUCCUGUCGAGUGCCAAUUAGGACCAGAAAAUCUA (.((((((((............)))))))).)........(((((((.(((((....)))))(((....((((....)))).))).)))))))(.((.((.....))))).......... (-29.28 = -31.28 + 2.00)

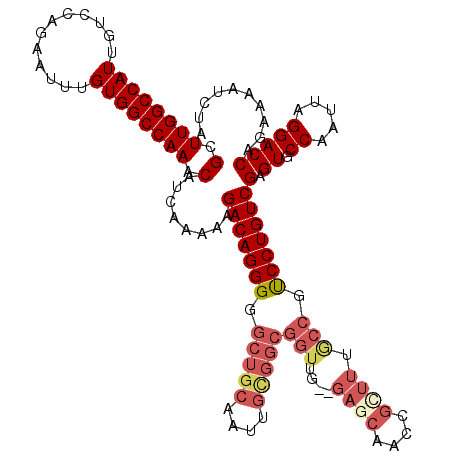

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:33:10 2006