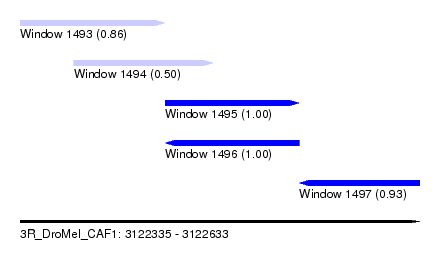
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,122,335 – 3,122,633 |
| Length | 298 |
| Max. P | 0.999779 |

| Location | 3,122,335 – 3,122,443 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -25.01 |
| Energy contribution | -23.68 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3122335 108 + 27905053 AAUACUAAUGCAAAUAAAUGUGAGUGCCAAUUGAAAUAGAAAACCGCGCUGCACACAAUCGAGGUGUCAAAGCGACGGCG------------AUGGUGAAUGUCGACGAGAGCGACAAAA ..(((((.(((.......(((((((((...((........))...))))).)))).........((((.....)))))))------------.)))))..(((((.(....))))))... ( -26.10) >DroPse_CAF1 9657 107 + 1 AAGACUAAUACAAAUAAAUGUGAGUGUCAAUUGAAAUAGAAAACCGCGUGCCACGCAAUCGAGGUGUCAAAGCGACGGCG------------ACGGCG-AUGCCGAGGAGACCGGCAAAA ..(((...((((......))))...)))...............(((((((((.((....)).)))(((.....))).)))------------.)))..-.(((((.(....))))))... ( -28.20) >DroSec_CAF1 9942 108 + 1 AAUACUAAUGCAAAUAAAUGUGAGUGCCAAUUGAAAUAGAAAACCGCGCUGCACACAAUCGAGGUGUCAAAGCGACGGCG------------AUGGUGAAUGUCGACGAGAGCGACAAAA ..(((((.(((.......(((((((((...((........))...))))).)))).........((((.....)))))))------------.)))))..(((((.(....))))))... ( -26.10) >DroSim_CAF1 15251 108 + 1 AAUACUAAUGCAAAUAAAUGUGAGUGCCAAUUGAAAUAGAAAACCGCGCUGCACACAAUCGAGGUGUCAAAGCGACGGCG------------AUGGUGAAUGUCGACGAGAGCGACAAAA ..(((((.(((.......(((((((((...((........))...))))).)))).........((((.....)))))))------------.)))))..(((((.(....))))))... ( -26.10) >DroPer_CAF1 9631 107 + 1 AGGACUAAUACAAAUAAAUGUGAGUGUCAAUUGAAAUAGAAAACCGCGUGCCACGCAAUCGAGGUGUCAAAGCGACGGCG------------ACGGCG-AUGCCGAGGAGACCGGCAAAA ..(((...((((......))))...)))...............(((((((((.((....)).)))(((.....))).)))------------.)))..-.(((((.(....))))))... ( -28.00) >DroYak_CAF1 13589 120 + 1 AAUACUAAUGCAAAUAAAUGUGAGUGCCAAUUGAAAUAGAAAACCGCGCUGCACACAAUCGAGGUGUCAAAGCGACGGCGGCGGCGACGGCGAUGGUGAAUGUCGACGAGAGCGACAAAA ...(((...(((......))).)))((((..............(((((((..((((.......))))...)))).)))((.((....)).)).))))...(((((.(....))))))... ( -33.40) >consensus AAUACUAAUGCAAAUAAAUGUGAGUGCCAAUUGAAAUAGAAAACCGCGCUGCACACAAUCGAGGUGUCAAAGCGACGGCG____________AUGGUGAAUGUCGACGAGAGCGACAAAA ...(((...(((......))).)))((((..............(((((((..((((.......))))...)))).)))...............))))...(((((.(....))))))... (-25.01 = -23.68 + -1.33)
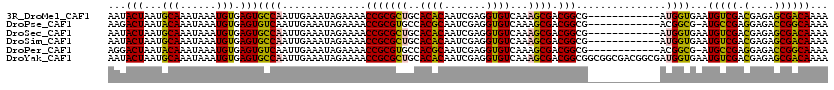
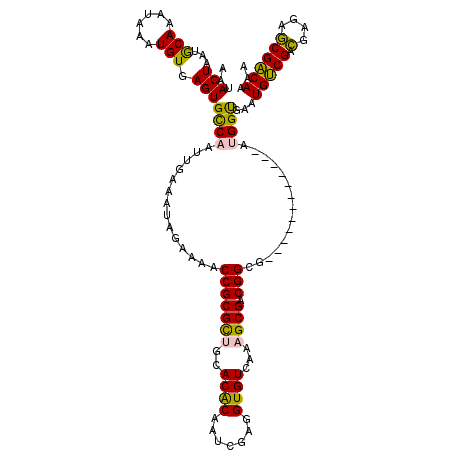
| Location | 3,122,375 – 3,122,479 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 94.55 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3122375 104 + 27905053 AAACCGCGCUGCACACAAUCGAGGUGUCAAAGCGACGGCG------------AUGGUGAAUGUCGACGAGAGCGACAAAAGUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAAC ......((((((((((.((((...((((.....)))).))------------)).))...(((((.(....))))))...))))))))..((((((.((((...)))))))))).. ( -35.30) >DroSec_CAF1 9982 104 + 1 AAACCGCGCUGCACACAAUCGAGGUGUCAAAGCGACGGCG------------AUGGUGAAUGUCGACGAGAGCGACAAAAGUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAAC ......((((((((((.((((...((((.....)))).))------------)).))...(((((.(....))))))...))))))))..((((((.((((...)))))))))).. ( -35.30) >DroSim_CAF1 15291 104 + 1 AAACCGCGCUGCACACAAUCGAGGUGUCAAAGCGACGGCG------------AUGGUGAAUGUCGACGAGAGCGACAAAAGUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAAC ......((((((((((.((((...((((.....)))).))------------)).))...(((((.(....))))))...))))))))..((((((.((((...)))))))))).. ( -35.30) >DroYak_CAF1 13629 116 + 1 AAACCGCGCUGCACACAAUCGAGGUGUCAAAGCGACGGCGGCGGCGACGGCGAUGGUGAAUGUCGACGAGAGCGACAAAAGUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAAC ......((((((((((.(((..(.((((.....)))).)(.((....)).)))).))...(((((.(....))))))...))))))))..((((((.((((...)))))))))).. ( -40.30) >consensus AAACCGCGCUGCACACAAUCGAGGUGUCAAAGCGACGGCG____________AUGGUGAAUGUCGACGAGAGCGACAAAAGUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAAC ......((((((((........(.((((.....)))).).....................(((((.(....))))))...))))))))..((((((.((((...)))))))))).. (-33.00 = -33.00 + 0.00)

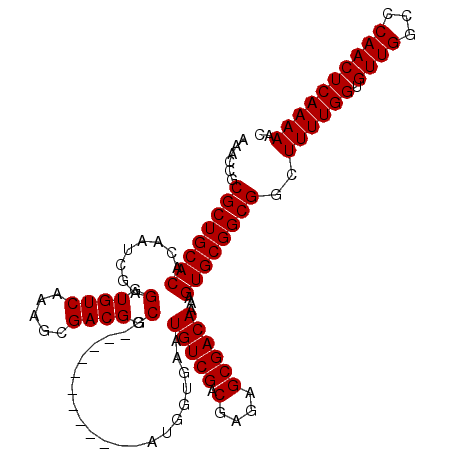
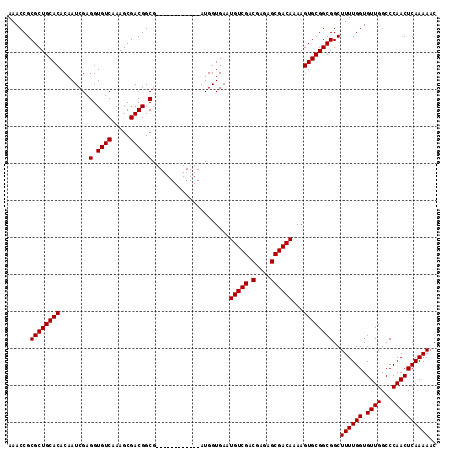
| Location | 3,122,443 – 3,122,543 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -27.16 |
| Energy contribution | -26.92 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3122443 100 + 27905053 GUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAACGUGAAAACGCAAAAAAUCGGAGCAAAACAAAGCAGGCCAAAACGCAACUAAAAAAGUGUUGCAU ((((((((.(((((.(((((((((...........(((....)))............((........)).)))))..))))......))))))))))))) ( -27.30) >DroSec_CAF1 10050 100 + 1 GUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAACGUGAAAACGCAAAAAAUCGGAGCAAAACAAAGCAGGCCAAAACGCAACUAAAAAAGUGUUGCAU ((((((((.(((((.(((((((((...........(((....)))............((........)).)))))..))))......))))))))))))) ( -27.30) >DroSim_CAF1 15359 100 + 1 GUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAACGUGAAAACGCAAAAAAUCGGAGCAAAACAAAGCAGGCCAAAACGCAACUAAAAAAGUGUUGCAU ((((((((.(((((.(((((((((...........(((....)))............((........)).)))))..))))......))))))))))))) ( -27.30) >DroEre_CAF1 9766 100 + 1 GUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAACGUGGAAACGCAAAAAAUCGGAGCAAAACAAAGCAGGCCAAAACACAACUAAAAAAGUGUUGCAU ((((((((.(((((.(((((((((............(((....)))...........((........)).)))))..))))......))))))))))))) ( -30.60) >DroYak_CAF1 13709 100 + 1 GUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAACGUGAAAACGCAAAAAAUCGGAGCAAAACAAAGCAGGCCAAAACACAACUAAAAAAGUGUUGCAU ((((((((.(((((.(((((((((...........(((....)))............((........)).)))))..))))......))))))))))))) ( -27.70) >consensus GUGCGGCGGCUUUUGGUGUUGGCCCAACUCAAAAACGUGAAAACGCAAAAAAUCGGAGCAAAACAAAGCAGGCCAAAACGCAACUAAAAAAGUGUUGCAU ((((((((.(((((.(((((((((............(((....)))...........((........)).)))))..))))......))))))))))))) (-27.16 = -26.92 + -0.24)

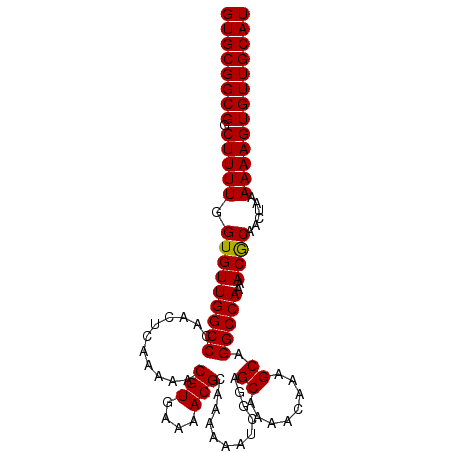
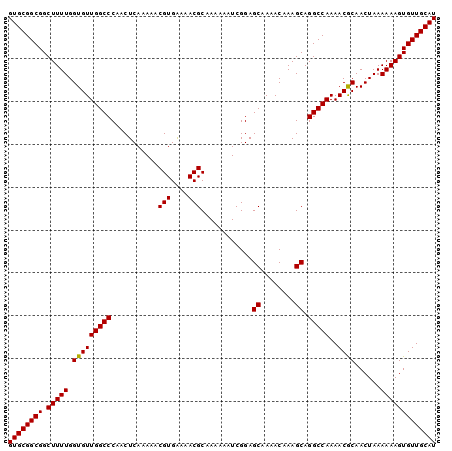
| Location | 3,122,443 – 3,122,543 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -27.70 |
| Energy contribution | -27.46 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3122443 100 - 27905053 AUGCAACACUUUUUUAGUUGCGUUUUGGCCUGCUUUGUUUUGCUCCGAUUUUUUGCGUUUUCACGUUUUUGAGUUGGGCCAACACCAAAAGCCGCCGCAC (((((((.........)))))))...(((..(((((((.(((..(((((((...((((....))))....)))))))..))).))..))))).))).... ( -28.50) >DroSec_CAF1 10050 100 - 1 AUGCAACACUUUUUUAGUUGCGUUUUGGCCUGCUUUGUUUUGCUCCGAUUUUUUGCGUUUUCACGUUUUUGAGUUGGGCCAACACCAAAAGCCGCCGCAC (((((((.........)))))))...(((..(((((((.(((..(((((((...((((....))))....)))))))..))).))..))))).))).... ( -28.50) >DroSim_CAF1 15359 100 - 1 AUGCAACACUUUUUUAGUUGCGUUUUGGCCUGCUUUGUUUUGCUCCGAUUUUUUGCGUUUUCACGUUUUUGAGUUGGGCCAACACCAAAAGCCGCCGCAC (((((((.........)))))))...(((..(((((((.(((..(((((((...((((....))))....)))))))..))).))..))))).))).... ( -28.50) >DroEre_CAF1 9766 100 - 1 AUGCAACACUUUUUUAGUUGUGUUUUGGCCUGCUUUGUUUUGCUCCGAUUUUUUGCGUUUCCACGUUUUUGAGUUGGGCCAACACCAAAAGCCGCCGCAC .((((((((..........)))))..(((..(((((((.(((..(((((((...((((....))))....)))))))..))).))..))))).)))))). ( -28.40) >DroYak_CAF1 13709 100 - 1 AUGCAACACUUUUUUAGUUGUGUUUUGGCCUGCUUUGUUUUGCUCCGAUUUUUUGCGUUUUCACGUUUUUGAGUUGGGCCAACACCAAAAGCCGCCGCAC .((((((((..........)))))..(((..(((((((.(((..(((((((...((((....))))....)))))))..))).))..))))).)))))). ( -28.40) >consensus AUGCAACACUUUUUUAGUUGCGUUUUGGCCUGCUUUGUUUUGCUCCGAUUUUUUGCGUUUUCACGUUUUUGAGUUGGGCCAACACCAAAAGCCGCCGCAC (((((((.........)))))))...(((..(((((((.(((..(((((((...((((....))))....)))))))..))).))..))))).))).... (-27.70 = -27.46 + -0.24)

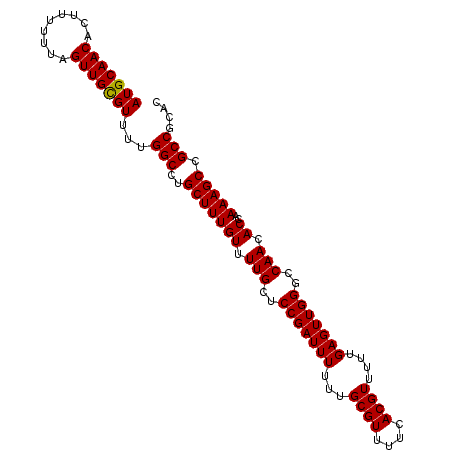
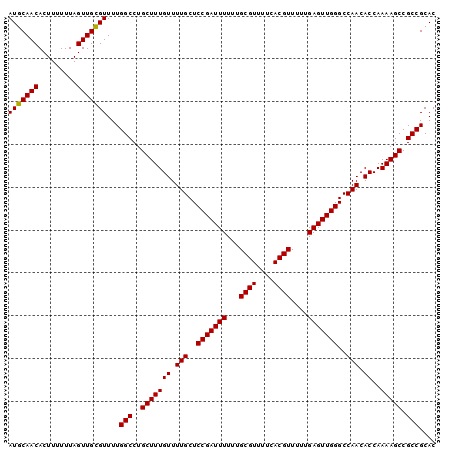
| Location | 3,122,543 – 3,122,633 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 98.52 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -26.03 |
| Energy contribution | -26.03 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3122543 90 - 27905053 AAAGCAACAAAGCUAACGCUCAGCGGUUCACACCCACGCACACCCGCAUGUGUGUGGGCCUAUCACACAGUGGCCAACUCCCCUUGCGCU ...((((...(((....)))....(((.(((.((((((((((......))))))))))...........))))))........))))... ( -27.00) >DroSec_CAF1 10150 90 - 1 AAAGCAACAAAGCUAAAGCUCAGCGGUUCACACCCACGCACACCCGCAUGUGUGUGGGCCUAUCACACAGUGGCCAACUCCCCUUGCGCU ..(((......)))..(((.(((.((........((((((((......))))))))((((...........)))).....)).))).))) ( -26.60) >DroSim_CAF1 15459 90 - 1 AAAGCAACAAAGCUAACGCUCAGCGGUUCACACCCACGCACUCCCGCAUGUGUGUGGGCCUAUCACACAGUGGCCAACUCCCCUUGCGCU ...((((...(((....)))....(((.(((.(((((((((........)))))))))...........))))))........))))... ( -26.60) >consensus AAAGCAACAAAGCUAACGCUCAGCGGUUCACACCCACGCACACCCGCAUGUGUGUGGGCCUAUCACACAGUGGCCAACUCCCCUUGCGCU ..(((.....(((....)))..(((((((((((.((.((......)).)).)))))))))..((((...))))............))))) (-26.03 = -26.03 + 0.00)

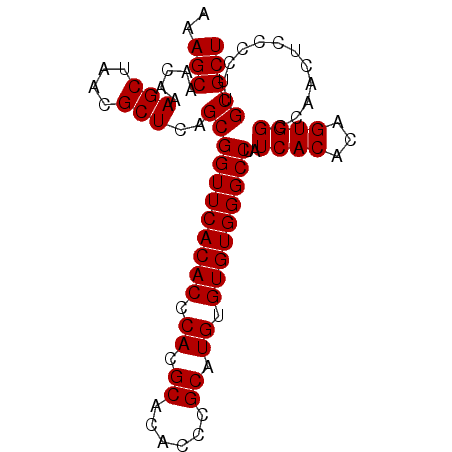
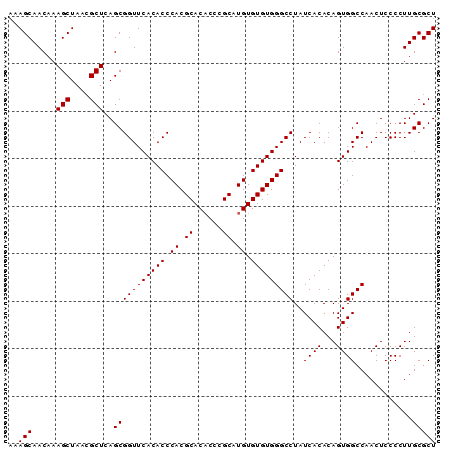
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:02 2006