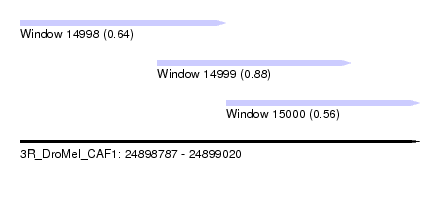
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,898,787 – 24,899,020 |
| Length | 233 |
| Max. P | 0.876187 |

| Location | 24,898,787 – 24,898,907 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -26.52 |
| Energy contribution | -27.56 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24898787 120 + 27905053 AUCUUGCGGAUAAUAACACAUUGCUUAUCUGUCGCCUGAGAUCUAUUCUCUUGAAGACAGCUGUAUGCCUAGAAGGAUAACACACGCUGUUUUGUUACUUUGGCAGUACAGGUAGCAUCA .....................((((.((((((((((.((((.....))))...(((((((((((.(.((.....))...).))).))))))))........))).).))))))))))... ( -31.10) >DroSec_CAF1 11790 120 + 1 AUCUUGCGGAUAAUAAAACAUUGCUAAUCUGUCGUCUGAGAUCUAUUCUCUUGAAGACAGAUGUAUGCCUAGAAGGAUAACACACGCUGUUUUGUUGCUUUGGCAGUACAGGUAGCAUCA .....................((((((((((((.((.((((.....))))..)).)))))))(((((((..(((((((((.((.....)).))))).)))))))).)))...)))))... ( -31.70) >DroSim_CAF1 11245 120 + 1 AUCUUGCGGAUAAUAACACAUUGCUAAUCUGUCGCCUGAGAUCUAUCCUCUUGAAGACAGCUGUAUGCCUAGAAGGAUAACACACGCUGUUUGGUUGCUUUGGCAGUACAGGUAGCAUCA .....................(((((.(((((((((.(((......((......((((((((((.(.((.....))...).))).)))))))))...))).))).).))))))))))... ( -28.00) >DroEre_CAF1 11852 120 + 1 AUCUUGUGGAUAAUAAUACAUUGCUAAUCUGUCGCUUAAGAUCUAUUCUCUUGAAGACAGCUGUAUGCCUAGAAGGAUAACACACGCUGUUUUGUUGCUUUGGCAGUACAGGUAGCAUCC ....((((........)))).(((((..(((((..((((((.......)))))).)))))(((((((((..(((((((((.((.....)).))))).)))))))).))))).)))))... ( -31.30) >DroYak_CAF1 11978 119 + 1 AUU-UUAGGGUAAUAAUACAUUGCUAAUCUGUCGCUUAAGAUCUAUUCUGUUGAAGACAGCUGUAUGCCUAGAAGGAUAACACACGCUGUUUUGUUGUUCUGGCAGUACAGGUAGCAUCC ...-...(((((....)))..(((((..(((((.....(((.....)))......)))))(((((((((.((((.(((((.((.....)).))))).)))))))).))))).))))).)) ( -33.70) >consensus AUCUUGCGGAUAAUAACACAUUGCUAAUCUGUCGCCUGAGAUCUAUUCUCUUGAAGACAGCUGUAUGCCUAGAAGGAUAACACACGCUGUUUUGUUGCUUUGGCAGUACAGGUAGCAUCA .....................(((((..(((((....((((.....)))).....)))))(((((((((.(((..(((((.((.....)).)))))..))))))).))))).)))))... (-26.52 = -27.56 + 1.04)



| Location | 24,898,867 – 24,898,980 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.66 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -30.60 |
| Energy contribution | -30.24 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24898867 113 + 27905053 CACACGCUGUUUUGUUACUUUGGCAGUACAGGUAGCAUCAAUAUCCCAUCUACUUUUUGGUAUGUCUGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCC .....((((...((((((((.(......)))))))))........(((((((((........((((.....))))((((((.....))))))...))))))...))).)))). ( -32.70) >DroSec_CAF1 11870 113 + 1 CACACGCUGUUUUGUUGCUUUGGCAGUACAGGUAGCAUCAAUAUCCCAUCUACUUUUUGGUCCGUCUGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCC .....((((...((((((((.(......)))))))))........(((((((((.....(((((.....))))).((((((.....))))))...))))))...))).)))). ( -34.90) >DroSim_CAF1 11325 113 + 1 CACACGCUGUUUGGUUGCUUUGGCAGUACAGGUAGCAUCAAUAUCCCAUCUACUUUUUGGUCCGUCGGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCC .....((((..((..(((....)))...))..)))).........(((((((((.....(((((.....))))).((((((.....))))))...))))))...)))...... ( -34.40) >DroEre_CAF1 11932 113 + 1 CACACGCUGUUUUGUUGCUUUGGCAGUACAGGUAGCAUCCAUAUCCCAUCUACUCUUUGGUCCGUCUGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCC .....((((...((((((((.(......)))))))))........(((((((((((...(((((.....))))).((((((.....)))))))).))))))...))).)))). ( -35.60) >DroYak_CAF1 12057 113 + 1 CACACGCUGUUUUGUUGUUCUGGCAGUACAGGUAGCAUCCAUAUCCCAUCUAGUUUUUGGUUCUUCUGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCC ...((.((((.(((((.....))))).)))))).......................((((((.((((((((....((((((.....))))))..))))))))...)))))).. ( -30.90) >consensus CACACGCUGUUUUGUUGCUUUGGCAGUACAGGUAGCAUCAAUAUCCCAUCUACUUUUUGGUCCGUCUGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCC .....((((....(((((.(((......)))))))).........(((((((((.....((((.......)))).((((((.....))))))...))))))...))).)))). (-30.60 = -30.24 + -0.36)

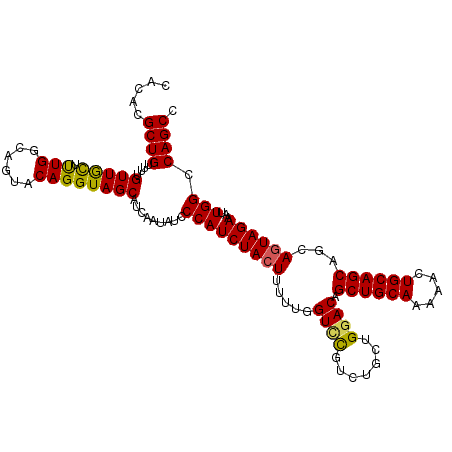
| Location | 24,898,907 – 24,899,020 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.66 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -25.62 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24898907 113 + 27905053 AUAUCCCAUCUACUUUUUGGUAUGUCUGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCCACUUACCAACGCUCUUCUCUUCAGUUCGAGUUCCUUUGAG ................((((((.((..(((((.(.((((((.....))))))..((((....)))))))))).)).)))))).((((..((....))...))))......... ( -28.00) >DroSec_CAF1 11910 113 + 1 AUAUCCCAUCUACUUUUUGGUCCGUCUGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCCACUUACCAACGCUCUCCUCUUCAGUUCGAGUUCCUUUGAG ................((((((..(((((((....((((((.....))))))..)))))))....))))))...((((.....((((.............)))).....)))) ( -27.42) >DroSim_CAF1 11365 113 + 1 AUAUCCCAUCUACUUUUUGGUCCGUCGGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCCACUUACCAACGCUCUUCUCUUCAGUUCGAGUUCCUUUGAG ................(((((..((.((((((.(.((((((.....))))))..((((....)))))))))))))..))))).((((..((....))...))))......... ( -30.60) >DroEre_CAF1 11972 113 + 1 AUAUCCCAUCUACUCUUUGGUCCGUCUGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCCACUUACCUACGCUCUUUACUUCAGUUCGAGUUGCUUUGAG ............(((..(((((..(((((((....((((((.....))))))..)))))))....)))))((.((((....((.............))..)))).))...))) ( -29.32) >DroYak_CAF1 12097 113 + 1 AUAUCCCAUCUAGUUUUUGGUUCUUCUGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCCACUUACCAACGCUCUUCACUUCAGUUCGAGUUCCUUUGAG ................(((((..((((((((....((((((.....))))))..))))))))..(((....)))...))))).((((...((....))..))))......... ( -27.00) >consensus AUAUCCCAUCUACUUUUUGGUCCGUCUGCUGGACAGCUGCAAAAACUGCAGCAGCAGUAGAAAUUGGCCAGCCACUUACCAACGCUCUUCUCUUCAGUUCGAGUUCCUUUGAG ................((((((..(((((((....((((((.....))))))..)))))))....))))))...((((.....((((.............)))).....)))) (-25.62 = -25.90 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:32:57 2006