
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,896,400 – 24,896,544 |
| Length | 144 |
| Max. P | 0.939879 |

| Location | 24,896,400 – 24,896,510 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24896400 110 - 27905053 CCGGGGGAUUCCCCGCUCCGCCCGUGGA-UCGACCAAUGUGAAUGCGAAUGUCAAGUGGCAUUGUACCAUGCUCAAAU--------GCACAAAUGCAAAAUGCAGAGUGCACACAUGAG .(((((....))))).((((....))))-.......(((((......((((((....))))))((((..(((.....(--------(((....))))....)))..)))).)))))... ( -33.10) >DroSec_CAF1 9381 110 - 1 CCGGGGGUUUCCCCGCUCCGCCCGUGGA-UCGGCCAAUGUGAAUGCGAAUGUCAAGUGGCAUUGUACCAUGCUCAAAU--------GCACAAAUGCAAAAUGCAGAGUGCACACAUGAG .(((((....)))))(((.(.(((....-.))).).(((((......((((((....))))))((((..(((.....(--------(((....))))....)))..)))).)))))))) ( -34.00) >DroSim_CAF1 8854 110 - 1 CCGGGGGAUUCCCCGCUCCGCCCGUGGA-UCGGCCAAUGUGAAUGCGAAUGUCAAGUGGCAUUGUACCAUGCUCAAAU--------GCACAAAUGCAAAAUGCAGAGUGCACACAUGAG .(((((....)))))(((.(.(((....-.))).).(((((......((((((....))))))((((..(((.....(--------(((....))))....)))..)))).)))))))) ( -34.00) >DroEre_CAF1 9445 107 - 1 CCAGGGGAUUCCCCGCUC---CCAUGGA-UUGGCCAAUGUGAAUGCGAAUGUCAAGUAGCAUUGUACAAUGCACAAAU--------GCACAAAUGCAAAAUGCAGAGUGCACACAUGAA (((.((((........))---)).))).-.......((((((((((............)))))((((..((((....(--------(((....))))...))))..)))).)))))... ( -29.90) >DroYak_CAF1 9586 116 - 1 CCAGGGGAUUCCCCGCUC---CCGUGGAAUCGGCCAAUGUGAAUGCGAAUGUCAAGUGGCAUUGUACAAUGCACAAAUGCACAAAUGCACAAAUGCAAAAUGCAGAGUGCACACAUGAG (((.((((........))---)).))).........(((((..(((.((((((....)))))))))...(((((...((((....((((....))))...))))..))))))))))... ( -36.40) >consensus CCGGGGGAUUCCCCGCUCCGCCCGUGGA_UCGGCCAAUGUGAAUGCGAAUGUCAAGUGGCAUUGUACCAUGCUCAAAU________GCACAAAUGCAAAAUGCAGAGUGCACACAUGAG ...((((...))))..........(((......)))(((((..(((.((((((....)))))))))....................((((...(((.....)))..)))).)))))... (-27.50 = -27.70 + 0.20)

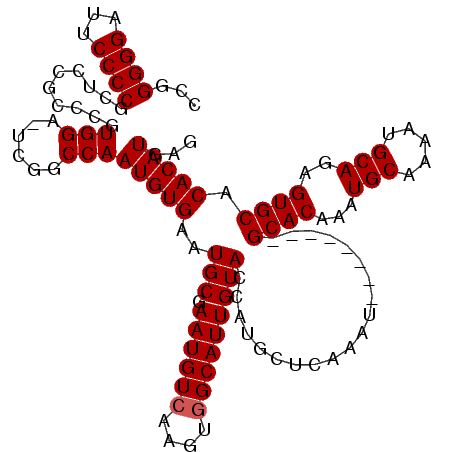

| Location | 24,896,433 – 24,896,544 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -30.24 |
| Energy contribution | -30.56 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24896433 111 + 27905053 -AUUUGAGCAUGGUACAAUGCCACUUGACAUUCGCAUUCACAUUGGUCGA-UCCACGGGCGGAGCGGGGAAUCCCCCGGUUGUUAAUGGGUUAAUGGUCUUUGAAUGUUCAAC -..((((((((....(((.((((.(((((..(((.(((......))))))-.(((..(((((..(((((....))))).)))))..))))))))))))..))).)))))))). ( -33.90) >DroSec_CAF1 9414 111 + 1 -AUUUGAGCAUGGUACAAUGCCACUUGACAUUCGCAUUCACAUUGGCCGA-UCCACGGGCGGAGCGGGGAAACCCCCGGUUGUUAAUGGGUUAAUGGUCUUUGAAUGUUCAAC -...((.((((......)))))).((((((((((......((((((((.(-(..((((.(((...(((....)))))).))))..)).)))))))).....)))))).)))). ( -38.70) >DroSim_CAF1 8887 111 + 1 -AUUUGAGCAUGGUACAAUGCCACUUGACAUUCGCAUUCACAUUGGCCGA-UCCACGGGCGGAGCGGGGAAUCCCCCGGCUGUUAAUGGGUUAAUGGUCUUUGAAUGUUCAAC -..(((((((((((.....)))..................((..((((((-((((..(((((..(((((....))))).)))))..))))))...))))..)).)))))))). ( -39.40) >DroEre_CAF1 9478 108 + 1 -AUUUGUGCAUUGUACAAUGCUACUUGACAUUCGCAUUCACAUUGGCCAA-UCCAUGG---GAGCGGGGAAUCCCCUGGUUGUUAAUGGGUUAAUGGUCUUUGAAUGUUCAAC -....(((((((....))))).))((((.....(((((((....((((((-(((((..---((.(((((....))))).))....))))))...)))))..))))))))))). ( -33.00) >DroYak_CAF1 9626 110 + 1 CAUUUGUGCAUUGUACAAUGCCACUUGACAUUCGCAUUCACAUUGGCCGAUUCCACGG---GAGCGGGGAAUCCCCUGGUUGUUAAUGGGUUAAUGGUCUUUGAAUGUUCAAC ....((.(((((....))))))).((((((((((......((((((((.((((((.((---((........)))).))).....))).)))))))).....)))))).)))). ( -32.70) >consensus _AUUUGAGCAUGGUACAAUGCCACUUGACAUUCGCAUUCACAUUGGCCGA_UCCACGGGCGGAGCGGGGAAUCCCCCGGUUGUUAAUGGGUUAAUGGUCUUUGAAUGUUCAAC ....((.((((......)))))).((((.....(((((((....(((((..((((..(((((..(((((....))))).)))))..))))....)))))..))))))))))). (-30.24 = -30.56 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:32:51 2006