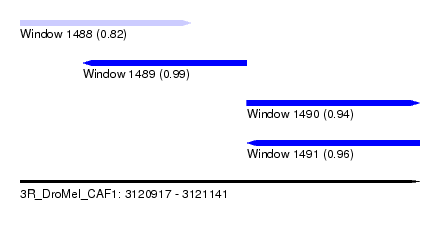
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,120,917 – 3,121,141 |
| Length | 224 |
| Max. P | 0.985034 |

| Location | 3,120,917 – 3,121,013 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.81 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -14.96 |
| Energy contribution | -15.07 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
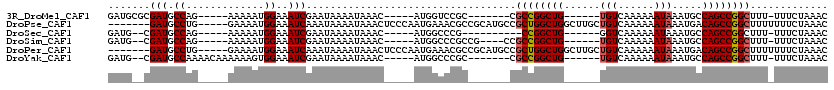
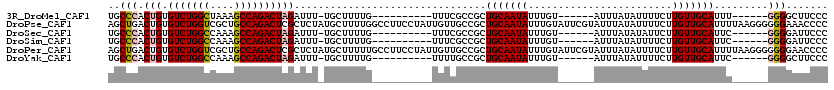
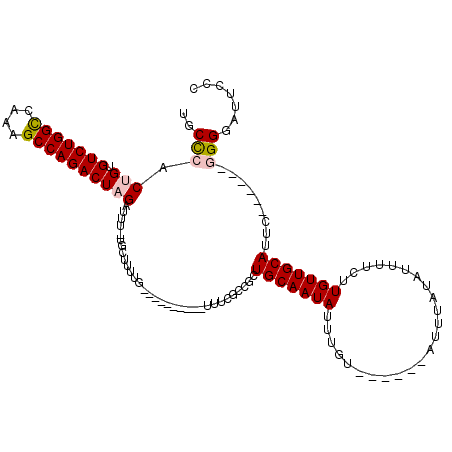
>3R_DroMel_CAF1 3120917 96 + 27905053 GAUGCGCGAUGCCAG-----AAAAAUGGAAAUCGAAUAAAAUAAAC-----AUGGUCCGC-------CGCCGGCUG------UGUCAAAAAAUAAAUGCCAGCCGGCUUU-UUUCUAAAC ......((((.(((.-----.....)))..))))............-----..((....)-------)((((((((------.((............))))))))))...-......... ( -24.20) >DroPse_CAF1 8124 108 + 1 -------GAUGCCUG-----GAAAAUGGAAAUCAAAUAAAAUAAACUCCCAAUGAAACGCCGCAUGCCGCUGGCUGGCUUGCUGUCAAAAAAUAAAUGACAGCCGGCUUUUUUUCUAAAC -------......((-----(((((.................................(((((.....)).))).((((.(((((((.........))))))).))))..)))))))... ( -24.90) >DroSec_CAF1 8548 91 + 1 GAUG--CGAUGCCAG-----AAAAAUGGAAAUCGAAUAAAAUAAAC-----AUGGCCCG----------CCGGCUG------GGUCAAAAAAUAAAUGCCAGCCGGCUUU-UUUCUAAAC ....--((((.(((.-----.....)))..))))............-----.......(----------(((((((------(...............)))))))))...-......... ( -25.06) >DroSim_CAF1 13821 97 + 1 GAUG--CGAUGCCAG-----AAAAAUGGAAAUCGAAUAAAAUAAAC-----AUGGCCCGCCG----CCGCCGGCUG------UGUCAAAAAAUAAAUGCCAGCCGGCUUU-UUUCUAAAC ....--((((.(((.-----.....)))..))))............-----..(((.....)----))((((((((------.((............))))))))))...-......... ( -26.30) >DroPer_CAF1 8086 108 + 1 -------GAUGCCUG-----GAAAAUGGAAAUCAAAUAAAAUAAACUCCCAAUGAAACGCCGCAUGCCGCUGGCUGGCUUGCUGUCAAAAAAUAAAUGACAGCCGGCUUUUUUUCUAAAC -------......((-----(((((.................................(((((.....)).))).((((.(((((((.........))))))).))))..)))))))... ( -24.90) >DroYak_CAF1 12041 99 + 1 GAUG--CGAUGCCAAAACAAAAAAGUGGAAAUCGAAUAAAAUAAAC-----AUGGCCCGC-------CGCCGGCUG------UGUCAAAAAAUAAAUGCCAGCCGGCUUU-UUUCUAAAC ...(--((..((((...........(.(....).)...........-----.)))).)))-------.((((((((------.((............))))))))))...-......... ( -23.30) >consensus GAUG__CGAUGCCAG_____AAAAAUGGAAAUCGAAUAAAAUAAAC_____AUGGCCCGC_______CGCCGGCUG______UGUCAAAAAAUAAAUGCCAGCCGGCUUU_UUUCUAAAC .......(((.((.............))..)))...................................((((((((......(((......))).....))))))))............. (-14.96 = -15.07 + 0.11)
| Location | 3,120,952 – 3,121,044 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -35.06 |
| Consensus MFE | -25.92 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.26 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3120952 92 - 27905053 CGGCCCCGCCCGACCUAAGCAUAUGGAAAUAGUUUAGAAAAAAGCCGGCUGGCAUUUAUUUUUUGACACAGCCGGCG---GCGGACCAUGUUUAU .((..(((((....((((((.(((....)))))))))......((((((((.((.........))...)))))))))---)))).))........ ( -37.50) >DroSec_CAF1 8581 84 - 1 CGCC-----CCGACCUAAGCAUAUGGAAAUAGUUUAGAAAAAAGCCGGCUGGCAUUUAUUUUUUGACCCAGCCGG------CGGGCCAUGUUUAU .(((-----(....((((((.(((....)))))))))......(((((((((...(((.....))).))))))))------)))))......... ( -34.00) >DroSim_CAF1 13854 90 - 1 CGCC-----CCGACCUAAGCAUAUGGAAAUAGUUUAGAAAAAAGCCGGCUGGCAUUUAUUUUUUGACACAGCCGGCGGCGGCGGGCCAUGUUUAU ((((-----((...((((((.(((....)))))))))......((((((((.((.........))...)))))))))).))))............ ( -35.60) >DroEre_CAF1 8215 85 - 1 CGC-------CGAACUAAGCAUAUGGAAAUAGUUUAGAAAAAAGCCGGCUGGCAUUUAUUUUUUGACACAGCCGGCG---GCGGGCCAUGUUUAU (((-------(...((((((.(((....)))))))))......((((((((.((.........))...)))))))))---)))............ ( -33.00) >DroYak_CAF1 12079 86 - 1 CGC------CCGACCUAAGCAUAUGGAAAUAGUUUAGAAAAAAGCCGGCUGGCAUUUAUUUUUUGACACAGCCGGCG---GCGGGCCAUGUUUAU .((------(((.(((((((.(((....)))))))).......((((((((.((.........))...)))))))))---))))))......... ( -35.20) >consensus CGCC_____CCGACCUAAGCAUAUGGAAAUAGUUUAGAAAAAAGCCGGCUGGCAUUUAUUUUUUGACACAGCCGGCG___GCGGGCCAUGUUUAU .........(((..((((((.(((....)))))))))......((((((((.((.........))...)))))))).....)))........... (-25.92 = -26.32 + 0.40)

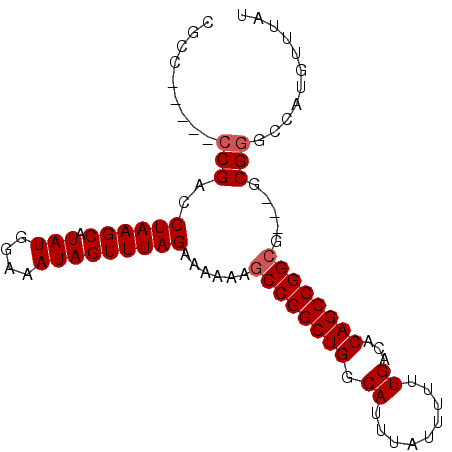
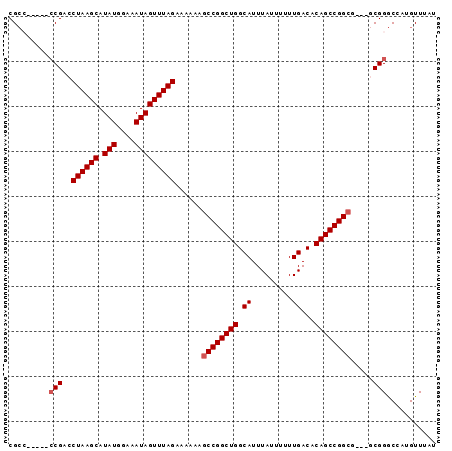
| Location | 3,121,044 – 3,121,141 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -19.93 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.939996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3121044 97 + 27905053 GGGAAGCCCC------AAAUGCAACAAGAAAAUAUAAAU------ACAAAUAUUGCAGCGGCGAAA----------CAAAAGCA-AAAUCUAGUCUGGCUUUAGCCAGACACAGUGGGCA .....(((((------...(((........(((((....------....)))))((....))....----------.....)))-.......(((((((....)))))))...).)))). ( -26.30) >DroPse_CAF1 8267 120 + 1 GGGGUUUCCCCCCUUAAAAUGCAACAAGAAAAUAUAAAUACGAAUACAAAUAUUGCAGCGGCAACAAUAGGAAGGCCAAAAGCAUAGAGCGAGUCUGGCAGCGACCAGACACAGUCAGCU ((((.....))))......(((((............................)))))(((....)........(((.....((.....))..((((((......))))))...))).)). ( -24.69) >DroSec_CAF1 8665 97 + 1 GGGAAUCCCC------GAAUGCAACAAGAAUAUAUAAAU------ACAAAUAUUGCAGCGGCGAAA----------CAAAAGCA-AAAUCUAGUCUGGCUUUGGCCAGACACAGUGGGCA (((....)))------...(((.((..............------.......((((....))))..----------........-.......(((((((....)))))))...))..))) ( -23.90) >DroSim_CAF1 13944 97 + 1 GGGAAUCCCC------GAAUGCAACAAGAAAAUAUAAAU------ACAAAUAUUGCAGCGGCGAAA----------CAAAAGCA-AAAUCUAGUCUGGCUUUGGCCAGACACAGUGGGCA (((....)))------...(((.((..............------.......((((....))))..----------........-.......(((((((....)))))))...))..))) ( -23.90) >DroPer_CAF1 8229 120 + 1 GGGGUUCCCCCCCUUAAAAUGCAACAAGAAAAUAUAAAUACGAAUACAAAUAUUGCAGCGGCAACAAUAGGAAGGCAAAAAGCAUAGAGCGAGUCUGGCAGCGACCAGACACAGUCAGCU ((((.....))))......(((((............................)))))(((....)........(((.....((.....))..((((((......))))))...))).)). ( -25.59) >DroYak_CAF1 12165 97 + 1 GGGAAGCCCC------GAAUGCAACAAGAAAAUAUAAAU------ACAAAUAUUGCAGCGGCAAAA----------CAAAAGCA-AAAUCUAGUCUGGCUUUGGCCAGACACAGUGGGCA .....(((((------...(((........(((((....------....)))))((....))....----------.....)))-.......(((((((....)))))))...).)))). ( -26.40) >consensus GGGAAUCCCC______AAAUGCAACAAGAAAAUAUAAAU______ACAAAUAUUGCAGCGGCAAAA__________CAAAAGCA_AAAUCUAGUCUGGCUUUGGCCAGACACAGUGGGCA (((....)))..........((.((...........................((((....))))............................(((((((....)))))))...))..)). (-19.93 = -20.02 + 0.08)

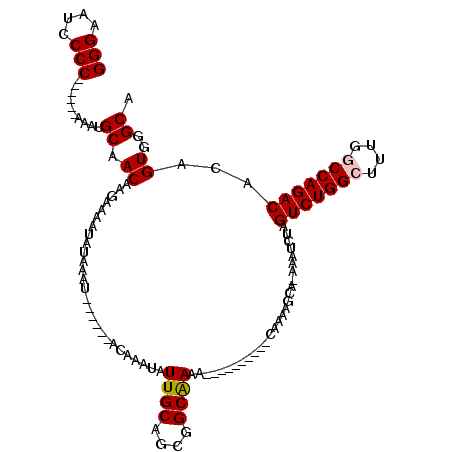
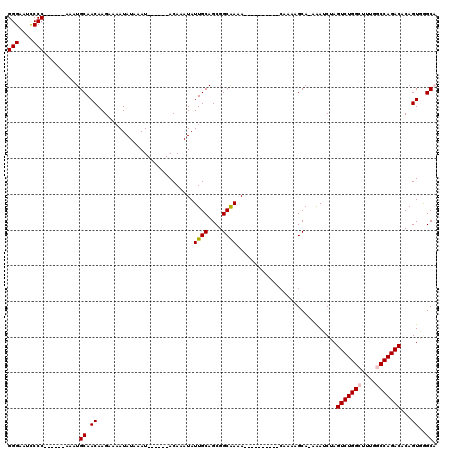
| Location | 3,121,044 – 3,121,141 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.81 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3121044 97 - 27905053 UGCCCACUGUGUCUGGCUAAAGCCAGACUAGAUUU-UGCUUUUG----------UUUCGCCGCUGCAAUAUUUGU------AUUUAUAUUUUCUUGUUGCAUUU------GGGGCUUCCC .((((.(((.(((((((....))))))))))....-........----------.........(((((((...((------(....))).....)))))))...------.))))..... ( -28.10) >DroPse_CAF1 8267 120 - 1 AGCUGACUGUGUCUGGUCGCUGCCAGACUCGCUCUAUGCUUUUGGCCUUCCUAUUGUUGCCGCUGCAAUAUUUGUAUUCGUAUUUAUAUUUUCUUGUUGCAUUUUAAGGGGGGAAACCCC (((.((.((.(((((((....))))))).)).))...)))...(((............)))..(((((((...((((........)))).....))))))).......((((....)))) ( -34.20) >DroSec_CAF1 8665 97 - 1 UGCCCACUGUGUCUGGCCAAAGCCAGACUAGAUUU-UGCUUUUG----------UUUCGCCGCUGCAAUAUUUGU------AUUUAUAUAUUCUUGUUGCAUUC------GGGGAUUCCC ......(((.(((((((....))))))))))....-.......(----------..((.(((.(((((((...((------((....))))...)))))))..)------)).))..).. ( -29.40) >DroSim_CAF1 13944 97 - 1 UGCCCACUGUGUCUGGCCAAAGCCAGACUAGAUUU-UGCUUUUG----------UUUCGCCGCUGCAAUAUUUGU------AUUUAUAUUUUCUUGUUGCAUUC------GGGGAUUCCC ......(((.(((((((....))))))))))....-.......(----------..((.(((.(((((((...((------(....))).....)))))))..)------)).))..).. ( -26.50) >DroPer_CAF1 8229 120 - 1 AGCUGACUGUGUCUGGUCGCUGCCAGACUCGCUCUAUGCUUUUUGCCUUCCUAUUGUUGCCGCUGCAAUAUUUGUAUUCGUAUUUAUAUUUUCUUGUUGCAUUUUAAGGGGGGGAACCCC (((.((.((.(((((((....))))))).)).))...)))..((.(((((((.....(((.((...(((((..(((....)))..))))).....)).))).....))))))).)).... ( -31.50) >DroYak_CAF1 12165 97 - 1 UGCCCACUGUGUCUGGCCAAAGCCAGACUAGAUUU-UGCUUUUG----------UUUUGCCGCUGCAAUAUUUGU------AUUUAUAUUUUCUUGUUGCAUUC------GGGGCUUCCC .((((.(((.(((((((....))))))))))....-........----------......((.(((((((...((------(....))).....)))))))..)------)))))..... ( -28.20) >consensus UGCCCACUGUGUCUGGCCAAAGCCAGACUAGAUUU_UGCUUUUG__________UUUCGCCGCUGCAAUAUUUGU______AUUUAUAUUUUCUUGUUGCAUUC______GGGGAUUCCC ..(((.(((.(((((((....))))))))))................................(((((((........................))))))).........)))....... (-19.59 = -19.81 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:56 2006