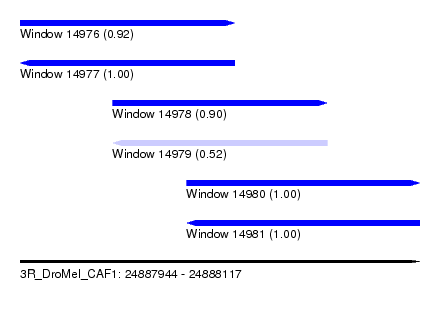
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,887,944 – 24,888,117 |
| Length | 173 |
| Max. P | 0.999825 |

| Location | 24,887,944 – 24,888,037 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.91 |
| Mean single sequence MFE | -45.33 |
| Consensus MFE | -24.83 |
| Energy contribution | -25.31 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24887944 93 + 27905053 UGCUGGAGCUCCCACUUCCGGAAGCAGCUGCAGCUGCCGCUGCUGAGCUUGUGGAGGCGGAUGUGGAGG--CUG------------------C-------AGCAGCGGCGGCUGAAGUGG .((....))..(((((((.....((....))(((((((((((((((((((.((.(......).)).)))--)).------------------)-------)))))))))))))))))))) ( -48.50) >DroVir_CAF1 1321 117 + 1 UAGCGGCGCCAACGCUAGUGGAAGCAGCAUUAGACGCCGCUGCCGAU---GUUGAGGCCUUUGCGGAGGAACUGCUUAAGGAUGCUUGAUUGCUGCUGGAGGCAGCGGCAUUCGAGGUGG (((((((((.((.(((.((....))))).)).).))))))))(((..---.......((((.((((.....))))..))))...(((((.((((((((....)))))))).))))).))) ( -46.50) >DroSec_CAF1 1020 93 + 1 UGCUGGAGCUCCCACUUCCGGAAGCAGCUGCAGCUGCCGCUGCUGAACUUGUGGAGGCGGAUGUGGAGG--CUG------------------C-------AGCAGCGGCGGCUGAUGUGG .((((.(((..((((.((((..((((((.((....)).))))))...(((...))).)))).))))..)--)).------------------)-------)))(((....)))....... ( -42.70) >DroSim_CAF1 537 93 + 1 UGCUGGAGCUCCCACUUCCGGAAGCAGCUGCAGCUGCCGCUGCUGAACUUGUGGAGGCGGAUGUGGAGG--CUG------------------C-------AGCAGCGGCGGCUGAAGUGG .((....))..(((((((.....((....))((((((((((((((..(((.((.(......).)).)))--...------------------)-------)))))))))))))))))))) ( -43.20) >DroEre_CAF1 1020 93 + 1 UGCUGGUGCUCCCACUUCCGGAAGCUACUGCAGCUGCCGCUGCUGAGCUUGUGGAGGCGGAUGUGGAGG--CUG------------------C-------AGCAGCGGCGGCUGAAGUGG .((....))..(((((((.....((....))(((((((((((((((((((.((.(......).)).)))--)).------------------)-------)))))))))))))))))))) ( -49.00) >DroMoj_CAF1 1029 117 + 1 UGGCGCCGCCAGCACUAGUGGACGCACCACCCGUUGCUGCCGAUGAU---GUUGAAGGCUUGGCCGAGGAACUGCUUGAGGGUACUUGACUGCUGAUCAAGGCAGCGGCAUUCGAGGUGG ..(((((((........)))).))).(((((((((((((((..((((---((..(..((((...((((......)))).))))..)..)).....)))).)))))))))....).))))) ( -42.10) >consensus UGCUGGAGCUCCCACUUCCGGAAGCAGCUGCAGCUGCCGCUGCUGAACUUGUGGAGGCGGAUGUGGAGG__CUG__________________C_______AGCAGCGGCGGCUGAAGUGG .......(((.((......)).))).....((((((((((((((........................................................))))))))))))))...... (-24.83 = -25.31 + 0.48)


| Location | 24,887,944 – 24,888,037 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.91 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -23.46 |
| Energy contribution | -24.61 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24887944 93 - 27905053 CCACUUCAGCCGCCGCUGCU-------G------------------CAG--CCUCCACAUCCGCCUCCACAAGCUCAGCAGCGGCAGCUGCAGCUGCUUCCGGAAGUGGGAGCUCCAGCA ((((((((((.(((((((((-------(------------------.((--(....................))))))))))))).)))((....)).....)))))))..((....)). ( -41.35) >DroVir_CAF1 1321 117 - 1 CCACCUCGAAUGCCGCUGCCUCCAGCAGCAAUCAAGCAUCCUUAAGCAGUUCCUCCGCAAAGGCCUCAAC---AUCGGCAGCGGCGUCUAAUGCUGCUUCCACUAGCGUUGGCGCCGCUA ...........((((((((.....)))))..........((((..((.(.....).)).)))).......---...)))(((((((.(((((((((.......)))))))))))))))). ( -41.40) >DroSec_CAF1 1020 93 - 1 CCACAUCAGCCGCCGCUGCU-------G------------------CAG--CCUCCACAUCCGCCUCCACAAGUUCAGCAGCGGCAGCUGCAGCUGCUUCCGGAAGUGGGAGCUCCAGCA ...........(((((((((-------(------------------.((--(....................))))))))))))).((((.((((.((..(....)..)))))).)))). ( -38.95) >DroSim_CAF1 537 93 - 1 CCACUUCAGCCGCCGCUGCU-------G------------------CAG--CCUCCACAUCCGCCUCCACAAGUUCAGCAGCGGCAGCUGCAGCUGCUUCCGGAAGUGGGAGCUCCAGCA ((((((((((.(((((((((-------(------------------.((--(....................))))))))))))).)))((....)).....)))))))..((....)). ( -38.95) >DroEre_CAF1 1020 93 - 1 CCACUUCAGCCGCCGCUGCU-------G------------------CAG--CCUCCACAUCCGCCUCCACAAGCUCAGCAGCGGCAGCUGCAGUAGCUUCCGGAAGUGGGAGCACCAGCA ..(((.((((.(((((((((-------(------------------.((--(....................))))))))))))).)))).))).(((((((....)))))))....... ( -42.65) >DroMoj_CAF1 1029 117 - 1 CCACCUCGAAUGCCGCUGCCUUGAUCAGCAGUCAAGUACCCUCAAGCAGUUCCUCGGCCAAGCCUUCAAC---AUCAUCGGCAGCAACGGGUGGUGCGUCCACUAGUGCUGGCGGCGCCA (((((.((..(((((((((((((((.....)))))).........))))).....(((...)))......---......))))....))))))).(((.((.(((....))).))))).. ( -36.80) >consensus CCACUUCAGCCGCCGCUGCU_______G__________________CAG__CCUCCACAUCCGCCUCCACAAGAUCAGCAGCGGCAGCUGCAGCUGCUUCCGGAAGUGGGAGCUCCAGCA ......((((.(((((((((........................................................))))))))).))))..((((((((((....)))))))...))). (-23.46 = -24.61 + 1.15)



| Location | 24,887,984 – 24,888,077 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 96.13 |
| Mean single sequence MFE | -40.36 |
| Consensus MFE | -36.30 |
| Energy contribution | -36.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24887984 93 + 27905053 UGCUGAGCUUGUGGAGGCGGAUGUGGAGGCUGCAGCAGCGGCGGCUGAAGUGGAGGCUCCAGCUGCGGCUGAGGUGGAGGCUCCUGACUUUCC ......((((....))))(((.((((((.((.((.((((.(((((((.(((....))).))))))).))))...)).)).))))..))..))) ( -40.50) >DroSec_CAF1 1060 93 + 1 UGCUGAACUUGUGGAGGCGGAUGUGGAGGCUGCAGCAGCGGCGGCUGAUGUGGAGGCUCCAGCUGCGGCUGAGGUGGAGGCUCCAGACUUUCC .(((...(....)..)))((((.(((((.((.((.((((.(((((((..((....))..))))))).))))...)).)).))))).)...))) ( -40.30) >DroSim_CAF1 577 93 + 1 UGCUGAACUUGUGGAGGCGGAUGUGGAGGCUGCAGCAGCGGCGGCUGAAGUGGAGGCUCCAGCUGCGGCUGAGGUGGAGGCUCCAGACUUUCC .(((...(....)..)))((((.(((((.((.((.((((.(((((((.(((....))).))))))).))))...)).)).))))).)...))) ( -41.60) >DroEre_CAF1 1060 93 + 1 UGCUGAGCUUGUGGAGGCGGAUGUGGAGGCUGCAGCAGCGGCGGCUGAAGUGGAGGCUCCAGCUGUGGCUGAGGUGGAGGCUCCUGACUUUCC ......((((....))))(((.((((((.((.((.((((.(((((((.(((....))).))))))).))))...)).)).))))..))..))) ( -39.20) >DroYak_CAF1 1065 93 + 1 UGCUGAGCUUGUGGAGGCGGAUGUGGAGGCUGCAGCAGCGGCGGCUGAUGUGGAGGCUCCAGCUGUGGCUGAGGUGGAACCUCCUGACUUCCC ......((((....))))(((.((((((((..(..((((.(((((((..((....))..))))))).))))..)..)..)))))..)).))). ( -40.20) >consensus UGCUGAGCUUGUGGAGGCGGAUGUGGAGGCUGCAGCAGCGGCGGCUGAAGUGGAGGCUCCAGCUGCGGCUGAGGUGGAGGCUCCUGACUUUCC .(((...(....)..)))(((.((((((.((.((.((((.(((((((.(((....))).))))))).))))...)).)).))))..)).))). (-36.30 = -36.50 + 0.20)



| Location | 24,887,984 – 24,888,077 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 96.13 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.98 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24887984 93 - 27905053 GGAAAGUCAGGAGCCUCCACCUCAGCCGCAGCUGGAGCCUCCACUUCAGCCGCCGCUGCUGCAGCCUCCACAUCCGCCUCCACAAGCUCAGCA (((..((..((((.((.((...((((.((.(((((((......))))))).)).)))).)).)).)))))).)))((........))...... ( -28.90) >DroSec_CAF1 1060 93 - 1 GGAAAGUCUGGAGCCUCCACCUCAGCCGCAGCUGGAGCCUCCACAUCAGCCGCCGCUGCUGCAGCCUCCACAUCCGCCUCCACAAGUUCAGCA ((...((.(((((.((.((...((((.((.((((..(......)..)))).)).)))).)).)).))))).))...))............... ( -25.80) >DroSim_CAF1 577 93 - 1 GGAAAGUCUGGAGCCUCCACCUCAGCCGCAGCUGGAGCCUCCACUUCAGCCGCCGCUGCUGCAGCCUCCACAUCCGCCUCCACAAGUUCAGCA ((...((.(((((.((.((...((((.((.(((((((......))))))).)).)))).)).)).))))).))...))............... ( -30.20) >DroEre_CAF1 1060 93 - 1 GGAAAGUCAGGAGCCUCCACCUCAGCCACAGCUGGAGCCUCCACUUCAGCCGCCGCUGCUGCAGCCUCCACAUCCGCCUCCACAAGCUCAGCA (((..((..((((.((.((...((((..(.(((((((......))))))).)..)))).)).)).)))))).)))((........))...... ( -24.60) >DroYak_CAF1 1065 93 - 1 GGGAAGUCAGGAGGUUCCACCUCAGCCACAGCUGGAGCCUCCACAUCAGCCGCCGCUGCUGCAGCCUCCACAUCCGCCUCCACAAGCUCAGCA .(((.((..((((((((((.((.......)).)))))))))).((.((((....)))).))..)).)))......((........))...... ( -28.30) >consensus GGAAAGUCAGGAGCCUCCACCUCAGCCGCAGCUGGAGCCUCCACUUCAGCCGCCGCUGCUGCAGCCUCCACAUCCGCCUCCACAAGCUCAGCA (((..((..((((.((.((...((((.((.(((((((......))))))).)).)))).)).)).)))))).))).................. (-24.50 = -24.98 + 0.48)


| Location | 24,888,016 – 24,888,117 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 92.87 |
| Mean single sequence MFE | -53.10 |
| Consensus MFE | -48.92 |
| Energy contribution | -49.68 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24888016 101 + 27905053 CAGCAGCGGCGGCUGAAGUGGAGGCUCCAGCUGCGGCUGAGGUGGAGGCUCCUGACUUUCCAGCCUGGCCGCUGCAGGAAGUAGCUGCUGCCGCCGCGGCG ..((.((((((((...((..(..(((((.((.((((((.(((((((((........)))))).))))))))).)).)).)))..)..)))))))))).)). ( -57.10) >DroSec_CAF1 1092 101 + 1 CAGCAGCGGCGGCUGAUGUGGAGGCUCCAGCUGCGGCUGAGGUGGAGGCUCCAGACUUUCCAGCCUGACCGCUGCAGGAAGAGGCUGCUGCCGCCGCGGCG ..((.((((((((...(.(((((.(((((.((.......)).))))).))))).).....((((((..((......))...))))))..)))))))).)). ( -54.40) >DroSim_CAF1 609 101 + 1 CAGCAGCGGCGGCUGAAGUGGAGGCUCCAGCUGCGGCUGAGGUGGAGGCUCCAGACUUUCCAGCCUGACCGCUGCAGGAAGAGGCUGCUGCCGCCGCGGCG ..((.((((((((.(((((((((.(((((.((.......)).))))).))))..))))).((((((..((......))...))))))..)))))))).)). ( -54.60) >DroEre_CAF1 1092 101 + 1 CAGCAGCGGCGGCUGAAGUGGAGGCUCCAGCUGUGGCUGAGGUGGAGGCUCCUGACUUUCCAGCUUGCCCGCUACAGGAUGAGGUUGCUGCCGCCGCAGCG ..((.((((((((.(((((((((.(((((.((.......)).))))).))))..))))).((((((..((......))...))))))..)))))))).)). ( -48.80) >DroYak_CAF1 1097 101 + 1 CAGCAGCGGCGGCUGAUGUGGAGGCUCCAGCUGUGGCUGAGGUGGAACCUCCUGACUUCCCAGCCUGGCCGCUGCAAGAAGAGGCUGCUGCCGCCGCAGCG ..(((((((((((((..(((((((.((((.((.......)).)))).)))))..))....)))))..))))))))........(((((.......))))). ( -50.60) >consensus CAGCAGCGGCGGCUGAAGUGGAGGCUCCAGCUGCGGCUGAGGUGGAGGCUCCUGACUUUCCAGCCUGACCGCUGCAGGAAGAGGCUGCUGCCGCCGCGGCG ..((.((((((((.(((((((((.(((((.((.......)).))))).))))..))))).((((((..((......))...))))))..)))))))).)). (-48.92 = -49.68 + 0.76)


| Location | 24,888,016 – 24,888,117 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.87 |
| Mean single sequence MFE | -49.14 |
| Consensus MFE | -44.82 |
| Energy contribution | -44.98 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24888016 101 - 27905053 CGCCGCGGCGGCAGCAGCUACUUCCUGCAGCGGCCAGGCUGGAAAGUCAGGAGCCUCCACCUCAGCCGCAGCUGGAGCCUCCACUUCAGCCGCCGCUGCUG ....((((((((.((((.......)))).(((((..((((....)))).((.....))......))))).(((((((......))))))).)))))))).. ( -49.20) >DroSec_CAF1 1092 101 - 1 CGCCGCGGCGGCAGCAGCCUCUUCCUGCAGCGGUCAGGCUGGAAAGUCUGGAGCCUCCACCUCAGCCGCAGCUGGAGCCUCCACAUCAGCCGCCGCUGCUG .((.((((((((..((((((...((......))..)))))).......(((((.(((((.(((....).)).))))).))))).....)))))))).)).. ( -49.90) >DroSim_CAF1 609 101 - 1 CGCCGCGGCGGCAGCAGCCUCUUCCUGCAGCGGUCAGGCUGGAAAGUCUGGAGCCUCCACCUCAGCCGCAGCUGGAGCCUCCACUUCAGCCGCCGCUGCUG ....((((((((.((((.......)))).(((((((((((....))))))(((.......))).))))).(((((((......))))))).)))))))).. ( -50.20) >DroEre_CAF1 1092 101 - 1 CGCUGCGGCGGCAGCAACCUCAUCCUGUAGCGGGCAAGCUGGAAAGUCAGGAGCCUCCACCUCAGCCACAGCUGGAGCCUCCACUUCAGCCGCCGCUGCUG .((.((((((((..........(((...(((......))))))......((((.(((((.((.......)).))))).))))......)))))))).)).. ( -42.70) >DroYak_CAF1 1097 101 - 1 CGCUGCGGCGGCAGCAGCCUCUUCUUGCAGCGGCCAGGCUGGGAAGUCAGGAGGUUCCACCUCAGCCACAGCUGGAGCCUCCACAUCAGCCGCCGCUGCUG .(((((....)))))...........((((((((..((((((...((..((((((((((.((.......)).)))))))))))).)))))))))))))).. ( -53.70) >consensus CGCCGCGGCGGCAGCAGCCUCUUCCUGCAGCGGCCAGGCUGGAAAGUCAGGAGCCUCCACCUCAGCCGCAGCUGGAGCCUCCACUUCAGCCGCCGCUGCUG .((.((((((((..((((((...((......))..))))))........((((.(((((.((.......)).))))).))))......)))))))).)).. (-44.82 = -44.98 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:32:39 2006