| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,886,056 – 24,886,162 |
| Length | 106 |
| Max. P | 0.631341 |

| Location | 24,886,056 – 24,886,162 |
|---|---|
| Length | 106 |
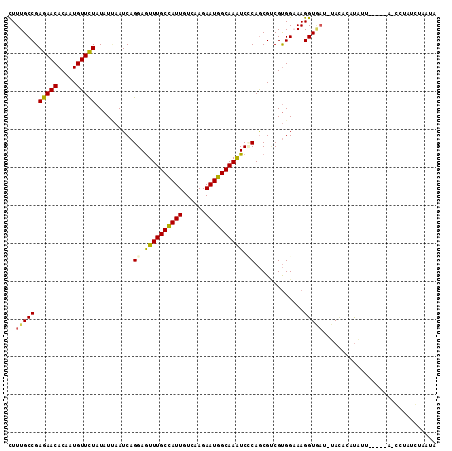
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -19.69 |
| Energy contribution | -19.33 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
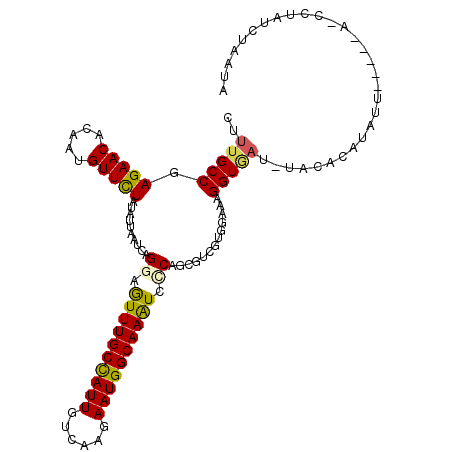
>3R_DroMel_CAF1 24886056 106 - 27905053 CUUUGCCGAGAACACAAUGUUCUAUAUUAAUCAGGAAUUUGCUAUUGUCAAGAAUGGCAAAUCCCAGCGUCGUGGAAAGGUGAU-UACGCAUAUU-----A-GCUAUCUAAUA ((((.((((((((.....))))...........((.((((((((((......)))))))))).))....))).).))))(((..-..))).((((-----(-(....)))))) ( -24.90) >DroVir_CAF1 387 111 - 1 UUUUGCCGAGAACACCAUGUUCUAUAUCAAUCAGGAGUUUGCCAUUGUCAAGAAUGGCAAAUCUCAACGGCGCGGCAAGGUAUGUUAUAUAUGUCG--AAACCAUCUGCUCUA ....(((((((((.....)))))...........((((((((((((......))))))))).)))..))))((((...(((...............--..)))..)))).... ( -31.83) >DroGri_CAF1 388 111 - 1 UUUUGCCGAGAACACCAUGUUUUAUAUCAAUCAGGAGUUUGCCAUUGUCAAGAAUGGCAAAUCACAGCGUCGUGGCAAGGUGAU-UAAUCACUUGCAUCGA-CCUAUGAACUA ..(((...(((((.....)))))....)))((((..((((((((((......))))))))))..)((.((((..(((((.(((.-...))))))))..)))-))).))).... ( -31.20) >DroSim_CAF1 358 106 - 1 CUUUGCCGAGAACACAAUGUUCUAUAUAAAUCAGGAGUUUGCUAUUGUCAAGAAUGGCAAGUCGCAGCGUCGUGGAAAGGUUAU-UACGCAUAUU-----A-GCUAUCUAAUA ...(((..(((((.....)))))...........((.(((((((((......))))))))))))))((((.((((.....))))-.)))).((((-----(-(....)))))) ( -23.20) >DroEre_CAF1 375 106 - 1 CUUUGCCGAGAACACAAUGUUCUAUAUUAAUCAGGAGUUUGCCAUUGUCAAGAAUGGCAAGUCCCAGCGCCGUGGAAAGGUGAA-UAUAAAUAUU-----A-CCUAUCUUAUA ....((..(((((.....)))))..........((..(((((((((......)))))))))..)).)).....(((.((((((.-........))-----)-))).))).... ( -27.80) >DroYak_CAF1 528 106 - 1 CUUUGCCGAGAACACAAUGUUCUAUAUUAAUCAGGAGUUUGCCAUUGUCAAGAAUGGCAAGUCUCAGCGCCGUGGAAAGGUAAU-UACUCAUGAU-----A-CCUCUGUUAUA ..(((((.(((((.....)))))...........((((((((((((......))))))))).))).............))))).-.....(((((-----(-....)))))). ( -25.20) >consensus CUUUGCCGAGAACACAAUGUUCUAUAUUAAUCAGGAGUUUGCCAUUGUCAAGAAUGGCAAAUCCCAGCGUCGUGGAAAGGUGAU_UACACAUAUU_____A_CCUAUCUAAUA ..(((((.(((((.....)))))..........((.((((((((((......)))))))))).)).............))))).............................. (-19.69 = -19.33 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:32:34 2006