
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,885,857 – 24,885,963 |
| Length | 106 |
| Max. P | 0.879345 |

| Location | 24,885,857 – 24,885,963 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.71 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -18.55 |
| Energy contribution | -19.47 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
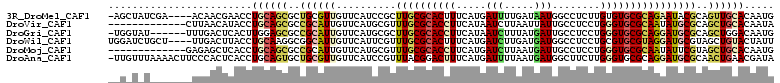
>3R_DroMel_CAF1 24885857 106 + 27905053 -AGCUAUCGA----ACAACGAACCUGCAGCGCUGCGUUGUUCAUCCGCUUGCGCACUUUCAUGAUUUUGAUAAUGGCCUCUUGUGUGCGCAGAAUACGCAGUUGCACAAUG -.((..(((.----....)))....)).(((((((((.((((.......(((((((...((.((..((......))..)).)).))))))))))))))))).)))...... ( -28.31) >DroVir_CAF1 193 98 + 1 -------------CUUAACAUACCUGCAGCGCCGCAUUGUUCAUGCGUUUGCGCACCUUCAUAAUCUUAAUUAUUGCCUCCUGGGUGCGCAAUAUGCGCAGCUGCACAAUA -------------...........(((((((((((((.....))))).((((((((((........................)))))))))).....)).))))))..... ( -31.66) >DroGri_CAF1 188 104 + 1 -UGGUAU------UUUGACUCACUUGGAGCGCCGCAUUGUUCAUGCGCUUGCGCACCUUCAUAAUCUUUAUGAUUGCCUCCUGGGUGCGCAGGAUGCGCAGCUGGACAAUG -.((..(------((..(.....)..)))..)).((((((((((((((((((((((((...(((((.....)))))......))))))))))...)))))..))))))))) ( -41.80) >DroWil_CAF1 177 107 + 1 UGGAUCUGCU----UUGACUUACCUGCAAGGCGGCAUUGUUCAUUCGUUUGCGCACUUUCAUGAUCUUGAUGAUGGCCUCCUGCGUGCGUAGGAUGCGUAGCUGUACUAUU ((((((((((----(((.(......)))))))))....)))))..((.(((((((...((((.((....)).))))..((((((....))))))))))))).))....... ( -29.10) >DroMoj_CAF1 194 98 + 1 -------------GAGAGCUCACCUGCAGCGCCGCAUUGUUCAUGCGUUUGCGCACCUUCAUGAUCUUAAUGAUUGCCUCCUGGGUGCGCAAUAUUCGUAGCUGCACAAUG -------------...........((((((..(((((.....))))).((((((((((....((((.....)))).......))))))))))........))))))..... ( -33.50) >DroAna_CAF1 175 110 + 1 -UUGUUUAAAACUUCCCACUCACCUGCAGUGCUGCGUUGUUCAUCCGUUUACGGACUUUCAUGAUUUUAAUGAUGGCUUCUUGGGUGCGCAGGAUGCGCAACUGAACGAUA -(((((((.............(((((.((.(((((((((.(((((((....))))......)))...))))).))))..)))))))((((.....))))...))))))).. ( -26.70) >consensus __G_U_U_______UUAACUCACCUGCAGCGCCGCAUUGUUCAUGCGUUUGCGCACCUUCAUGAUCUUAAUGAUGGCCUCCUGGGUGCGCAGGAUGCGCAGCUGCACAAUG ........................((((((..((((((..........((((((((((.....(((.....)))........))))))))))))))))..))))))..... (-18.55 = -19.47 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:32:33 2006