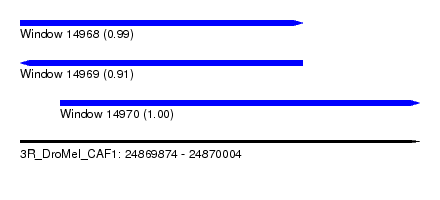
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,869,874 – 24,870,004 |
| Length | 130 |
| Max. P | 0.995303 |

| Location | 24,869,874 – 24,869,966 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.11 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -17.26 |
| Energy contribution | -16.82 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24869874 92 + 27905053 ---------------------------AAUCAUUGCAUUUAUUGAGCUUUGUAUCUUUUGAUAGAAAAUGUAGAUGGUUCAUUUUGUGUAUCAUGGCGAUCUUCUUUCUGAGCUCAUCG ---------------------------...............(((((((..((((....))))((((....((((.((.(((..........))))).))))..)))).)))))))... ( -16.10) >DroSec_CAF1 3138 118 + 1 UUCAUUUAUUGACUUUCAAAUGAAAAAGAGUAUUU-AUUUGUAGAGCUUUCUAUCUUUUGAUAGAAAAGAUAGAUGGCUCGUUUUGUGCAUCAUGGCGAUCUUCUUCCCGAGCUCAUAG (((((((..........)))))))...((((....-.......(((((.((((((((((.....)))))))))).))))).............(((.((......))))).)))).... ( -29.50) >DroSim_CAF1 3137 118 + 1 UUCAUUUAUUGACUUUCAAAUGAAAAAGAGUAUUG-AUUUGUAGAGCUUUCUAUCUUUUGAUAGAAAAGAUAGAUGGCUCGUUUUGUGCAUCAGGGCGAUCUUCUUUCCGAGCUCAUAG (((((((..........)))))))...((((.(((-.......(((((.((((((((((.....)))))))))).)))))((((((.....))))))...........))))))).... ( -31.10) >DroEre_CAF1 3196 101 + 1 ----------AACUUU-ACAUUAACA----AAUUC-AUUAGAUGGGCUUUCUAUCUUUUAAUAGAAAAGAUAGAAAGUUCGU--AGUGUAACAUGGCGAUCUUCUUCCCGAGCUCAUAG ----------....((-((((((...----...((-....))(((((((((((((((((.....))))))))))))))))))--)))))))..(((.((......)))))......... ( -26.70) >DroYak_CAF1 3311 90 + 1 ----------AACU----------------AAUUG-AUUUUGAGAGCUGUGUAUCUUUUGAUAGAAAAGAUAGAUAGUUCAU--AGUGCAUCAUGGCGAUCUUCCUUCCGAGCUCAUAG ----------....----------------...((-(.((((.(((((((.((((((((.....)))))))).)))))))..--..(((......)))..........)))).)))... ( -21.50) >consensus __________AACUUU_A_AU_AA_A__A_AAUUG_AUUUGUAGAGCUUUCUAUCUUUUGAUAGAAAAGAUAGAUGGUUCGUUUUGUGCAUCAUGGCGAUCUUCUUUCCGAGCUCAUAG ...........................................(((((.((((((((((.....)))))))))).))))).......((......))((.(((......))).)).... (-17.26 = -16.82 + -0.44)

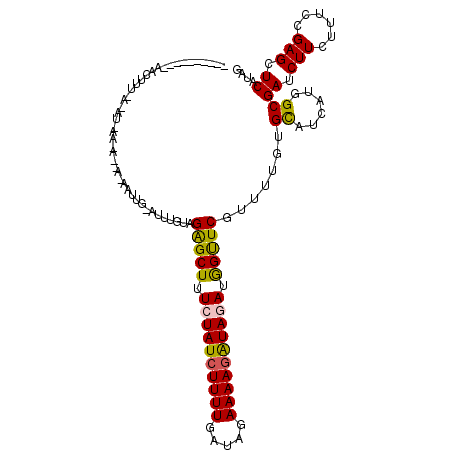
| Location | 24,869,874 – 24,869,966 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.11 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -11.37 |
| Energy contribution | -12.17 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24869874 92 - 27905053 CGAUGAGCUCAGAAAGAAGAUCGCCAUGAUACACAAAAUGAACCAUCUACAUUUUCUAUCAAAAGAUACAAAGCUCAAUAAAUGCAAUGAUU--------------------------- ...((((((.((((((.((((.(.(((..........)))..).))))...))))))(((....)))....))))))...............--------------------------- ( -11.40) >DroSec_CAF1 3138 118 - 1 CUAUGAGCUCGGGAAGAAGAUCGCCAUGAUGCACAAAACGAGCCAUCUAUCUUUUCUAUCAAAAGAUAGAAAGCUCUACAAAU-AAAUACUCUUUUUCAUUUGAAAGUCAAUAAAUGAA ...(((..(((.((((((((..((......)).......((((..((((((((((.....))))))))))..)))).......-......))))))))...)))...)))......... ( -26.00) >DroSim_CAF1 3137 118 - 1 CUAUGAGCUCGGAAAGAAGAUCGCCCUGAUGCACAAAACGAGCCAUCUAUCUUUUCUAUCAAAAGAUAGAAAGCUCUACAAAU-CAAUACUCUUUUUCAUUUGAAAGUCAAUAAAUGAA ...(((((((((...(.....)...)))).)).......((((..((((((((((.....))))))))))..))))......)-)).........((((((((........)))))))) ( -25.50) >DroEre_CAF1 3196 101 - 1 CUAUGAGCUCGGGAAGAAGAUCGCCAUGUUACACU--ACGAACUUUCUAUCUUUUCUAUUAAAAGAUAGAAAGCCCAUCUAAU-GAAUU----UGUUAAUGU-AAAGUU---------- .((((.((((........))..))))))(((((..--((((((((((((((((((.....)))))))))))))....((....-)).))----)))...)))-))....---------- ( -21.20) >DroYak_CAF1 3311 90 - 1 CUAUGAGCUCGGAAGGAAGAUCGCCAUGAUGCACU--AUGAACUAUCUAUCUUUUCUAUCAAAAGAUACACAGCUCUCAAAAU-CAAUU----------------AGUU---------- ....(((((.(((((((((((.(.((((......)--)))..).)))).)))))))((((....))))...))))).......-.....----------------....---------- ( -16.20) >consensus CUAUGAGCUCGGAAAGAAGAUCGCCAUGAUGCACAAAACGAACCAUCUAUCUUUUCUAUCAAAAGAUAGAAAGCUCUACAAAU_CAAUA_U__U_UU_AU_U_AAAGUC__________ ....(((((((........(((.....)))........)))....((((((((((.....))))))))))..))))........................................... (-11.37 = -12.17 + 0.80)

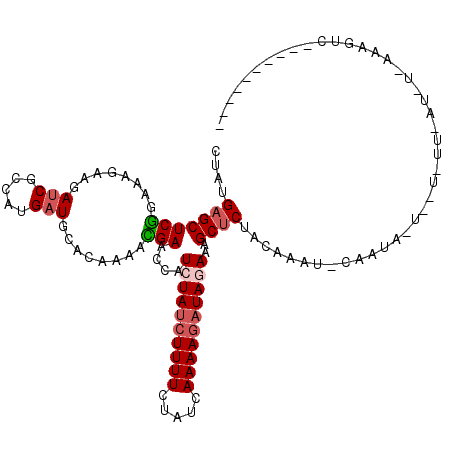
| Location | 24,869,887 – 24,870,004 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -35.56 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.02 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24869887 117 + 27905053 AUUGAGCUUUGUAUCUUUUGAUAGAAAAUGUAGAUGGUUCAUUUUGUGUAUCAUGGCGAUCUUCUUUCUGAGCUCAUCGAUUUUCAUAGAGUCUUGGAUUCAUUGGC--CUGCACUCUG ..((((((.(.(((.((((.....)))).))).).))))))....(((((....(((.(.......((..(((((.............))).))..)).....).))--)))))).... ( -23.22) >DroSec_CAF1 3177 117 + 1 GUAGAGCUUUCUAUCUUUUGAUAGAAAAGAUAGAUGGCUCGUUUUGUGCAUCAUGGCGAUCUUCUUCCCGAGCUCAUAGAUUUUCAUAGAGACUUGGAUUCACUGGC--CUGCACUCUG ...(((((.((((((((((.....)))))))))).))))).....(((((....(((((......))((((((((.((........))))).)))))........))--)))))).... ( -38.00) >DroSim_CAF1 3176 117 + 1 GUAGAGCUUUCUAUCUUUUGAUAGAAAAGAUAGAUGGCUCGUUUUGUGCAUCAGGGCGAUCUUCUUUCCGAGCUCAUAGAUUUUCAUAGAGUCUUGGAUUCACUGGC--CUGCACUCUG ...(((((.((((((((((.....)))))))))).))))).....(((((....(((.(.......(((((((((.((........))))).)))))).....).))--)))))).... ( -39.30) >DroEre_CAF1 3220 114 + 1 GAUGGGCUUUCUAUCUUUUAAUAGAAAAGAUAGAAAGUUCGU--AGUGUAACAUGGCGAUCUUCUUCCCGAGCUCAUAGAUA-UCAUUGAGUUUUGGAUUCUUUGGC--CUGCACUCUG .((((((((((((((((((.....))))))))))))))))))--((((((....(((.(......(((.(((((((......-....))))))).))).....).))--)))))))... ( -41.30) >DroYak_CAF1 3324 117 + 1 UGAGAGCUGUGUAUCUUUUGAUAGAAAAGAUAGAUAGUUCAU--AGUGCAUCAUGGCGAUCUUCCUUCCGAGCUCAUAGAUUUUCAUAGACUUUCGGAUUCUUUGGCCACUGCACUCUG ...(((((((.((((((((.....)))))))).)))))))..--((((((...((((.(.......((((((....((........))....)))))).....).)))).))))))... ( -36.00) >consensus GUAGAGCUUUCUAUCUUUUGAUAGAAAAGAUAGAUGGUUCGUUUUGUGCAUCAUGGCGAUCUUCUUUCCGAGCUCAUAGAUUUUCAUAGAGUCUUGGAUUCAUUGGC__CUGCACUCUG ...(((((.((((((((((.....)))))))))).))))).....(((((.....((.(.......(((((((((.............))).)))))).....).))...))))).... (-28.42 = -28.02 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:32:30 2006