| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
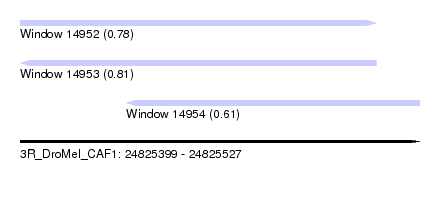
| Location | 24,825,399 – 24,825,527 |
| Length | 128 |
| Max. P | 0.811339 |

| Location | 24,825,399 – 24,825,513 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.94 |
| Mean single sequence MFE | -34.89 |
| Consensus MFE | -19.70 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
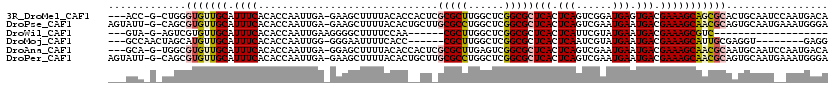
>3R_DroMel_CAF1 24825399 114 + 27905053 ---ACC-G-CUGGGUGUUGCAUUUCACACCAAUUGA-GAAGCUUUUACACCACUCGCGCUUGGCUCGGCGCUCACUCAGUCGGAUGAGUGACGAAAGCAGCGCACUGCAAUCCAAUGACA ---...-(-(.((((((.((.(((((.......)))-)).))....)))))....(((((..(((...((.(((((((......)))))))))..)))))))).).))............ ( -37.50) >DroPse_CAF1 37583 117 + 1 AGUAUU-G-CAGCGUGUUGCAUUUCACACCAAUUGA-GAAGCUUUUACACUGCUUGCGCCUGGCUCGGCGCUCACUCAGUCGAAUGAAUGACGAAAGCAACGCAGUGCAAUGAAAUGGGA ..((((-(-((..(((((((.((((........(((-(((((.........))))(((((......)))))...))))((((......)))))))))))))))..)))))))........ ( -40.50) >DroWil_CAF1 35010 90 + 1 ---GUA-G-AGUCGUGUUGCAUUUCACACCAAUUGAAGGGGCUUUUCCAA------CGCUUGGCUCGGCGCUCACUCAUUCGUAUGAAUGACGAAAGCGUC------------------- ---...-(-((((((((((.......(.((.......))).......)))------)))..)))))((((((..(((((((....)))))).)..))))))------------------- ( -26.54) >DroMoj_CAF1 71140 102 + 1 ---GCCAACUAGCAUGUUGCAUUUCACACCAAUUGG-GGGAAUUUUCACC------CGCUUGGCUCGGCGCUCACUCAAUCGUAUGAAUGACGAAAGCAUUGCGAGGU--------GAGG ---(((.....((.....))........((((.(((-(.((....)).))------)).))))...))).((((((...(((((...(((.(....))))))))))))--------))). ( -32.20) >DroAna_CAF1 36164 114 + 1 ---GCA-G-UGGCGUGUUGCAUUUCACACCAAUUGA-GGAGCUUUUACACCACUCGCGCUUGAGUCGGCGCUCACUCAGUCGAAUGAAUGACGAAAGCAACGCAAUGCAAUCCAAUGACA ---(((-.-....(((((((.((((........(((-(((((.......(((((((....))))).)).)))).))))((((......)))))))))))))))..)))............ ( -32.10) >DroPer_CAF1 38137 117 + 1 AGUAUU-G-CAGCGUGUUGCAUUUCACACCAAUUGA-GAAGCUUUUACACUGCUUGCGCCUGGCUCGGCGCUCACUCAGUCGAAUGAAUGACGAAAGCAACGCAGUGCAAUGAAAUGGGA ..((((-(-((..(((((((.((((........(((-(((((.........))))(((((......)))))...))))((((......)))))))))))))))..)))))))........ ( -40.50) >consensus ___ACA_G_CAGCGUGUUGCAUUUCACACCAAUUGA_GAAGCUUUUACAC__CU_GCGCUUGGCUCGGCGCUCACUCAGUCGAAUGAAUGACGAAAGCAACGCAAUGCAAU__AAUGAGA .............(((((((.((((..............................(((((......)))))(((.(((......))).))).)))))))))))................. (-19.70 = -20.28 + 0.58)
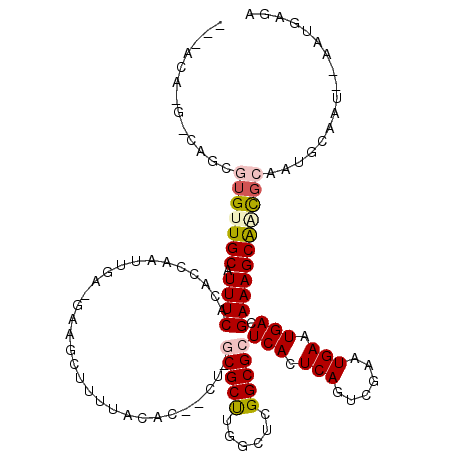
| Location | 24,825,399 – 24,825,513 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.94 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -20.50 |
| Energy contribution | -22.10 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24825399 114 - 27905053 UGUCAUUGGAUUGCAGUGCGCUGCUUUCGUCACUCAUCCGACUGAGUGAGCGCCGAGCCAAGCGCGAGUGGUGUAAAAGCUUC-UCAAUUGGUGUGAAAUGCAACACCCAG-C-GGU--- .........(((((...(((((((((.(((((((((......))))))).))..))))..)))))(((.(((......))).)-))....(((((........)))))..)-)-)))--- ( -37.80) >DroPse_CAF1 37583 117 - 1 UCCCAUUUCAUUGCACUGCGUUGCUUUCGUCAUUCAUUCGACUGAGUGAGCGCCGAGCCAGGCGCAAGCAGUGUAAAAGCUUC-UCAAUUGGUGUGAAAUGCAACACGCUG-C-AAUACU .........((((((.((.(((((((((.(((((((......)))))))(((((((...((((....))(((......))).)-)...))))))))))).))))).)).))-)-)))... ( -38.90) >DroWil_CAF1 35010 90 - 1 -------------------GACGCUUUCGUCAUUCAUACGAAUGAGUGAGCGCCGAGCCAAGCG------UUGGAAAAGCCCCUUCAAUUGGUGUGAAAUGCAACACGACU-C-UAC--- -------------------...((((((.((((((((....))))))))(((((((...(((.(------((.....)))..)))...))))))))))).)).........-.-...--- ( -22.50) >DroMoj_CAF1 71140 102 - 1 CCUC--------ACCUCGCAAUGCUUUCGUCAUUCAUACGAUUGAGUGAGCGCCGAGCCAAGCG------GGUGAAAAUUCCC-CCAAUUGGUGUGAAAUGCAACAUGCUAGUUGGC--- ..((--------(((.(((...((((.(((((((((......))))))).))..))))...)))------)))))........-((((((((((((........)))))))))))).--- ( -38.30) >DroAna_CAF1 36164 114 - 1 UGUCAUUGGAUUGCAUUGCGUUGCUUUCGUCAUUCAUUCGACUGAGUGAGCGCCGACUCAAGCGCGAGUGGUGUAAAAGCUCC-UCAAUUGGUGUGAAAUGCAACACGCCA-C-UGC--- .......(((..((.(((((..((((...(((((((......)))))))((((........))))))))..)))))..)))))-.....(((((((........)))))))-.-...--- ( -34.20) >DroPer_CAF1 38137 117 - 1 UCCCAUUUCAUUGCACUGCGUUGCUUUCGUCAUUCAUUCGACUGAGUGAGCGCCGAGCCAGGCGCAAGCAGUGUAAAAGCUUC-UCAAUUGGUGUGAAAUGCAACACGCUG-C-AAUACU .........((((((.((.(((((((((.(((((((......)))))))(((((((...((((....))(((......))).)-)...))))))))))).))))).)).))-)-)))... ( -38.90) >consensus UCUCAUU__AUUGCACUGCGUUGCUUUCGUCAUUCAUUCGACUGAGUGAGCGCCGAGCCAAGCGC_AG__GUGUAAAAGCUCC_UCAAUUGGUGUGAAAUGCAACACGCUG_C_AAC___ ..........(((((((((..(((((((((((((((......)))))))....)))...)))))...)))))))))..............((((((........)))))).......... (-20.50 = -22.10 + 1.60)


| Location | 24,825,433 – 24,825,527 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.04 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -25.50 |
| Energy contribution | -24.33 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24825433 94 - 27905053 ------------------------GAAUCCCCGC--CUUCUGUCAUUGGAUUGCAGUGCGCUGCUUUCGUCACUCAUCCGACUGAGUGAGCGCCGAGCCAAGCGCGAGUGGUGUAAAAGC ------------------------........((--..(((......)))..))..(((((..(((...(((((((......)))))))((((........)))))))..)))))..... ( -30.40) >DroPse_CAF1 37620 117 - 1 CAG-CACCGCACCGCCCAGCGCACC--UCUCUGCCUCGCCUCCCAUUUCAUUGCACUGCGUUGCUUUCGUCAUUCAUUCGACUGAGUGAGCGCCGAGCCAGGCGCAAGCAGUGUAAAAGC ..(-(...((........))(((..--....)))...))...........(((((((((..........(((((((......)))))))(((((......)))))..))))))))).... ( -36.30) >DroSec_CAF1 41340 94 - 1 ------------------------GAAUCCCCGC--CUUCUGUCAUUGGAUUGCAGUGCGCUGCUUUCGUCACUCAUCCGACUGAGUGAGCGCCAAGCCAAGCGCGAGUGGUGUAAAAGC ------------------------........((--..(((......)))..))..(((((..(((...(((((((......)))))))((((........)))))))..)))))..... ( -30.40) >DroSim_CAF1 43542 93 - 1 ------------------------G-AUCCCCGC--UUUCUGUCAUUGGAUUGCAGUGCGCUGCUUUCGUCACUCAUCCGACUGAGUGAGCGCCGAGCCAAGCGCGAGUGGUGUAAAAGC ------------------------.-....((((--((.(((.((......))))).(((((((((.(((((((((......))))))).))..))))..)))))))))))......... ( -31.60) >DroAna_CAF1 36198 117 - 1 CACCAACCCCACCGACCAACCCGUCACCCACCG---CGGCUGUCAUUGGAUUGCAUUGCGUUGCUUUCGUCAUUCAUUCGACUGAGUGAGCGCCGACUCAAGCGCGAGUGGUGUAAAAGC .............(((......)))...(((((---(.((.(((....))).)).(((((((....(((.(.(((((((....))))))).).)))....)))))))))))))....... ( -33.30) >DroPer_CAF1 38174 115 - 1 CAG-CACCGCACCG--CAGCGCACC--UCUCUGCCUCGCCUCCCAUUUCAUUGCACUGCGUUGCUUUCGUCAUUCAUUCGACUGAGUGAGCGCCGAGCCAGGCGCAAGCAGUGUAAAAGC ..(-(...((...(--(((.(....--.).))))...))...........(((((((((..........(((((((......)))))))(((((......)))))..)))))))))..)) ( -37.70) >consensus ________________________C_AUCCCCGC__CGUCUGUCAUUGGAUUGCAGUGCGCUGCUUUCGUCACUCAUCCGACUGAGUGAGCGCCGAGCCAAGCGCGAGUGGUGUAAAAGC ....................................................((..((((((((((...(((((((......)))))))((((........))))))))))))))...)) (-25.50 = -24.33 + -1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:32:16 2006