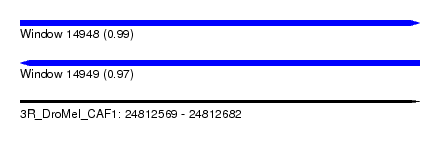
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,812,569 – 24,812,682 |
| Length | 113 |
| Max. P | 0.990575 |

| Location | 24,812,569 – 24,812,682 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
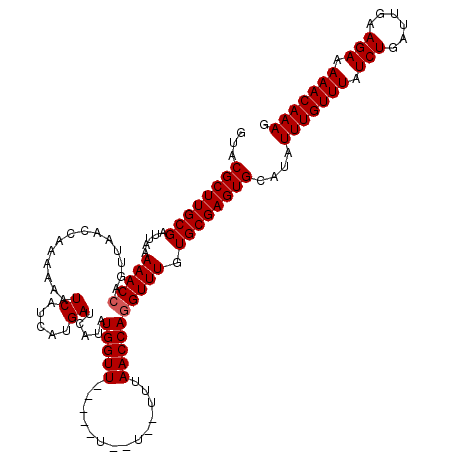
>3R_DroMel_CAF1 24812569 113 + 27905053 GUACGCUUGCGAUUAAAAACCAGUUAACCAAAAAAUCAUCAUGAUCAUAUGGUU-----UUUU--UUUAACCAGGUUUGUGCGAGUGCAUAUUUGUUUAUCUGAUUGAAGAAAAACAAAG ...((((((((.....(((((.(((((..((((((((((.((....))))))))-----))))--.)))))..))))).))))))))....(((((((.(((......))).))))))). ( -30.10) >DroSec_CAF1 28735 111 + 1 GUACGCUUGCGAUUAAAAACCAGUUAACCAAAAAAUCAUCAUGAUCAUAUGGUU-----U--U--UUUAACCAGGUUUGUGCGAGUGCAUAUUUGUUUAUCUGAUUGAAGAAAAACAAAG ...((((((((.....(((((.(((((...(((((((((.((....))))))))-----)--)--))))))..))))).))))))))....(((((((.(((......))).))))))). ( -28.70) >DroSim_CAF1 30655 111 + 1 GUACGCUUGCGAUUAAAAACCAGUUAACCCAAAAAUCAUCAUGAUCAUAUGGUU-----U--U--UUUAACCAGGUUUGUGCGAGUGCAUAUUUGUUUAUCUGAUUGAAGAAAAACAAAG ...((((((((.....(((((.(((((...(((((((((.((....))))))))-----)--)--))))))..))))).))))))))....(((((((.(((......))).))))))). ( -28.70) >DroEre_CAF1 23613 111 + 1 GUACGCUUGCGAUUAAAAACCAGCUAACCAAAAAAUCAUCAUGACCAAGUGGUU-----U--U--UUUAACCACGUUUAUGCGAGUGCAUAUUUGUUUAUCUGAUUGAAGAAAAACAAAG ...(((((((((((...................))))...........((((((-----.--.--...))))))......)))))))....(((((((.(((......))).))))))). ( -24.91) >DroYak_CAF1 24274 118 + 1 GUACGCUUGCGAUUAAAAACCAGCUAACCAAAAAAUCAUCAUGACCAAAUGGUUGAUUUU--UUUUGAAACCAGGUUUAUGCGAGUGCAUAUUUGUUUAUCUGAUUGAAGAAAAACAAAG ...((((((((.....(((((........(((((((((((((......)))).)))))))--)).........))))).))))))))....(((((((.(((......))).))))))). ( -27.73) >consensus GUACGCUUGCGAUUAAAAACCAGUUAACCAAAAAAUCAUCAUGAUCAUAUGGUU_____U__U__UUUAACCAGGUUUGUGCGAGUGCAUAUUUGUUUAUCUGAUUGAAGAAAAACAAAG ...((((((((.....(((((..............((.....)).....(((((..............)))))))))).))))))))....(((((((.(((......))).))))))). (-22.28 = -22.48 + 0.20)


| Location | 24,812,569 – 24,812,682 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -19.95 |
| Energy contribution | -19.87 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
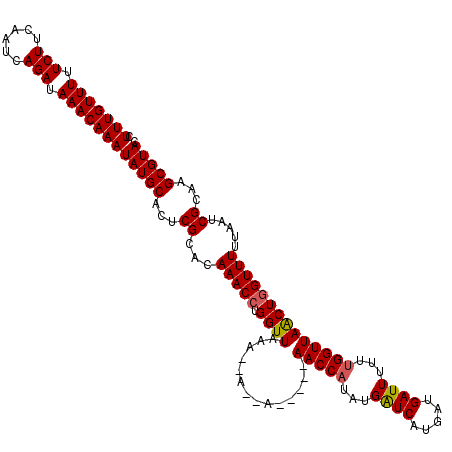
>3R_DroMel_CAF1 24812569 113 - 27905053 CUUUGUUUUUCUUCAAUCAGAUAAACAAAUAUGCACUCGCACAAACCUGGUUAAA--AAAA-----AACCAUAUGAUCAUGAUGAUUUUUUGGUUAACUGGUUUUUAAUCGCAAGCGUAC .(((((((.(((......))).)))))))(((((...((...(((((.(((((..--....-----.((((...((((.....))))...)))))))))))))).....))...))))). ( -22.41) >DroSec_CAF1 28735 111 - 1 CUUUGUUUUUCUUCAAUCAGAUAAACAAAUAUGCACUCGCACAAACCUGGUUAAA--A--A-----AACCAUAUGAUCAUGAUGAUUUUUUGGUUAACUGGUUUUUAAUCGCAAGCGUAC .(((((((.(((......))).)))))))(((((...((...(((((.(((((..--.--.-----.((((...((((.....))))...)))))))))))))).....))...))))). ( -23.00) >DroSim_CAF1 30655 111 - 1 CUUUGUUUUUCUUCAAUCAGAUAAACAAAUAUGCACUCGCACAAACCUGGUUAAA--A--A-----AACCAUAUGAUCAUGAUGAUUUUUGGGUUAACUGGUUUUUAAUCGCAAGCGUAC .(((((((.(((......))).)))))))(((((...((...(((((.((((((.--.--.-----..(((...((((.....))))..))).))))))))))).....))...))))). ( -23.90) >DroEre_CAF1 23613 111 - 1 CUUUGUUUUUCUUCAAUCAGAUAAACAAAUAUGCACUCGCAUAAACGUGGUUAAA--A--A-----AACCACUUGGUCAUGAUGAUUUUUUGGUUAGCUGGUUUUUAAUCGCAAGCGUAC .(((((((.(((......))).)))))))(((((....)))))...(((((((((--(--.-----.((((.(((..((.((.....)).))..))).))))))))))))))........ ( -26.90) >DroYak_CAF1 24274 118 - 1 CUUUGUUUUUCUUCAAUCAGAUAAACAAAUAUGCACUCGCAUAAACCUGGUUUCAAAA--AAAAUCAACCAUUUGGUCAUGAUGAUUUUUUGGUUAGCUGGUUUUUAAUCGCAAGCGUAC .(((((((.(((......))).)))))))(((((...((...(((((.((((((((((--((.(((((((....)))..))))..))))))))..))))))))).....))...))))). ( -26.10) >consensus CUUUGUUUUUCUUCAAUCAGAUAAACAAAUAUGCACUCGCACAAACCUGGUUAAA__A__A_____AACCAUAUGAUCAUGAUGAUUUUUUGGUUAACUGGUUUUUAAUCGCAAGCGUAC .(((((((.(((......))).)))))))(((((...((...(((((.((((..............(((((...((((.....))))...)))))))))))))).....))...))))). (-19.95 = -19.87 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:32:12 2006