
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,810,373 – 24,810,497 |
| Length | 124 |
| Max. P | 0.668532 |

| Location | 24,810,373 – 24,810,471 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -18.70 |
| Consensus MFE | -12.08 |
| Energy contribution | -12.42 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24810373 98 + 27905053 CGAAG-------UCCUGGCAAAAUGGUGAAACAUUAAAUU-UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGCCCAAAUA----AAAUGGAAAAUGG .....-------(((.(((..((((......)))).....-((((((((((........))))))))))....)))((....)).........----....)))...... ( -17.50) >DroVir_CAF1 55574 82 + 1 -------------------AAAAUGGUGAAACAUUAAAUU-UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGUCCAAAUACAUGCGAGU-------- -------------------....(((..............-((((((((((........))))))))))..((((.......))))))).............-------- ( -14.60) >DroPse_CAF1 23028 90 + 1 AGACG-------UGGGCGAGAAAUGGUGAAACAUUAAAUU-UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGCCCAAAUA----AAAUG-------- .....-------((((((...((((......)))).....-((((((((((........))))))))))..............))))))....----.....-------- ( -21.80) >DroGri_CAF1 56344 79 + 1 -------------------AAAAUGGUGAAACAUUAAAUUUUAUCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGUCCAAAUU----CGGCU-------- -------------------....((((((((.......))))))))......(((((....(((......)))((.......)))))))....----.....-------- ( -13.60) >DroAna_CAF1 20337 98 + 1 AGAGGGGCCUGCGCCUGGCAAAAUGGUAAAACAUUAAAUU-UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGCCCAAAUA----AAAUGG------- ....((((..(((........((((......)))).....-((((((((((........))))))))))...))).((....)))))).....----......------- ( -22.90) >DroPer_CAF1 23230 90 + 1 AGACG-------UGGGCGAGAAAUGGUGAAACAUUAAAUU-UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGCCCAAAUA----AAAUG-------- .....-------((((((...((((......)))).....-((((((((((........))))))))))..............))))))....----.....-------- ( -21.80) >consensus AGA_G_______U____G_AAAAUGGUGAAACAUUAAAUU_UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGCCCAAAUA____AAAUG________ .......................(((...............((((((((((........))))))))))...(((.......))).)))..................... (-12.08 = -12.42 + 0.33)



| Location | 24,810,373 – 24,810,471 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -17.66 |
| Consensus MFE | -11.97 |
| Energy contribution | -11.92 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24810373 98 - 27905053 CCAUUUUCCAUUU----UAUUUGGGCGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA-AAUUUAAUGUUUCACCAUUUUGCCAGGA-------CUUCG ......(((....----((..(((((((.((((((......)).))))...)))))))..))..((.((-((....(((......))))))).)))))-------..... ( -15.70) >DroVir_CAF1 55574 82 - 1 --------ACUCGCAUGUAUUUGGACGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA-AAUUUAAUGUUUCACCAUUUU------------------- --------....(((.(((..(((((((.((((((......)).))))...)))))))..))).)))..-.....................------------------- ( -18.80) >DroPse_CAF1 23028 90 - 1 --------CAUUU----UAUUUGGGCGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA-AAUUUAAUGUUUCACCAUUUCUCGCCCA-------CGUCU --------.....----....(((((((....))(((((((.......((((((......))))))...-....)))))))............)))))-------..... ( -18.14) >DroGri_CAF1 56344 79 - 1 --------AGCCG----AAUUUGGACGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGAUAAAAUUUAAUGUUUCACCAUUUU------------------- --------...((----(((.(((((((.((((((......)).))))...)))))))...))))).........................------------------- ( -15.20) >DroAna_CAF1 20337 98 - 1 -------CCAUUU----UAUUUGGGCGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA-AAUUUAAUGUUUUACCAUUUUGCCAGGCGCAGGCCCCUCU -------......----.....((((((....)).((.....((((......)))).........(((.-.....((((......))))......)))))..)))).... ( -20.00) >DroPer_CAF1 23230 90 - 1 --------CAUUU----UAUUUGGGCGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA-AAUUUAAUGUUUCACCAUUUCUCGCCCA-------CGUCU --------.....----....(((((((....))(((((((.......((((((......))))))...-....)))))))............)))))-------..... ( -18.14) >consensus ________CAUUU____UAUUUGGGCGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA_AAUUUAAUGUUUCACCAUUUU_C____A_______C_UCU .................((..(((((((.((((((......)).))))...)))))))..))................................................ (-11.97 = -11.92 + -0.05)

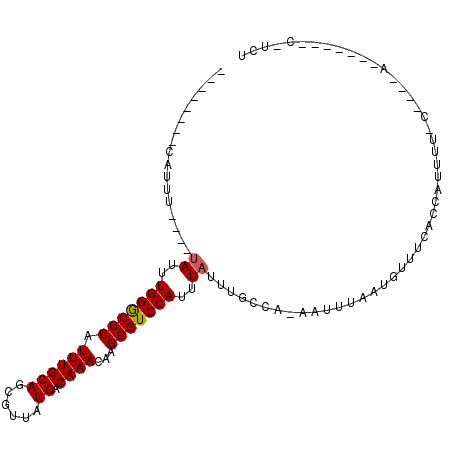

| Location | 24,810,406 – 24,810,497 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -12.90 |
| Consensus MFE | -9.05 |
| Energy contribution | -9.55 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24810406 91 + 27905053 -UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGCCCAAAUA----AAAUGGAAAAUGGA-AAAUGAAAAUGAAAAACC----AAAAAGG---- -((((((((((........))))))))))...(((.......))).(((....----...)))....(((.-..((....))......))----)......---- ( -13.20) >DroVir_CAF1 55595 87 + 1 -UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGUCCAAAUACAUGCGAGU--------A-AAAUAAAAAUGAAGGACC----AAAAAGG---- -((((((((((........))))))))))....((.......))((((...(((......))--------)-..((....))...)))).----.......---- ( -12.80) >DroPse_CAF1 23061 87 + 1 -UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGCCCAAAUA----AAAUG--------A-AAAUGAAAAUGGAAAACC----AAAAAAGUAAA -((((((((((........))))))))))...(((.......))).(((....----..((.--------.-..)).....)))......----........... ( -12.90) >DroGri_CAF1 56365 88 + 1 UUAUCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGUCCAAAUU----CGGCU--------A-AAAUAUAAAUGAAGACCAACAAAAAAAGG---- ...................(((((.(((...((((.......))))((.....----.))..--------.-.............))).))))).......---- ( -11.20) >DroMoj_CAF1 54424 84 + 1 -UGCCAAAUAAAUGGACACUUGUUUGUCAUAACGCUACAAAUGCGUCCAAGAA----CGACU--------AAAAAUAAAAAUGAAAAACC----GAAAGGG---- -...........(((((....(((......)))((.......)))))))....----.....--------..................((----....)).---- ( -14.40) >DroPer_CAF1 23263 87 + 1 -UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGCCCAAAUA----AAAUG--------A-AAAUGAAAAUGGAAAACC----AAAAACGUAAA -((((((((((........))))))))))...(((.......))).(((....----..((.--------.-..)).....)))......----........... ( -12.90) >consensus _UGGCAAAUAAAUGGACACUUGUUUGUCAUAACGCUGCAAAUGCGCCCAAAUA____AAAUG________A_AAAUAAAAAUGAAAAACC____AAAAAGG____ .((((((((((........))))))))))...(((.......)))............................................................ ( -9.05 = -9.55 + 0.50)

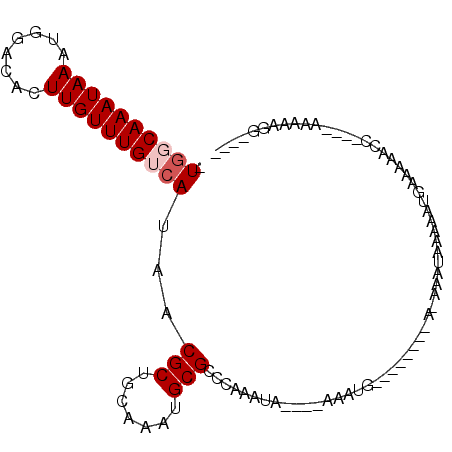
| Location | 24,810,406 – 24,810,497 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -14.88 |
| Consensus MFE | -11.85 |
| Energy contribution | -11.47 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24810406 91 - 27905053 ----CCUUUUU----GGUUUUUCAUUUUCAUUU-UCCAUUUUCCAUUU----UAUUUGGGCGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA- ----((.....----))................-.........((...----((..(((((((.((((((......)).))))...)))))))..))..))...- ( -12.70) >DroVir_CAF1 55595 87 - 1 ----CCUUUUU----GGUCCUUCAUUUUUAUUU-U--------ACUCGCAUGUAUUUGGACGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA- ----((.....----))................-.--------....(((.(((..(((((((.((((((......)).))))...)))))))..))).)))..- ( -19.10) >DroPse_CAF1 23061 87 - 1 UUUACUUUUUU----GGUUUUCCAUUUUCAUUU-U--------CAUUU----UAUUUGGGCGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA- ..........(----((....))).........-.--------.....----((..(((((((.((((((......)).))))...)))))))..)).......- ( -13.80) >DroGri_CAF1 56365 88 - 1 ----CCUUUUUUUGUUGGUCUUCAUUUAUAUUU-U--------AGCCG----AAUUUGGACGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGAUAA ----.............................-.--------...((----(((.(((((((.((((((......)).))))...)))))))...))))).... ( -15.20) >DroMoj_CAF1 54424 84 - 1 ----CCCUUUC----GGUUUUUCAUUUUUAUUUUU--------AGUCG----UUCUUGGACGCAUUUGUAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGGCA- ----.......----....................--------.((((----....(((((((.(((((.........)))))...)))))))......)))).- ( -14.60) >DroPer_CAF1 23263 87 - 1 UUUACGUUUUU----GGUUUUCCAUUUUCAUUU-U--------CAUUU----UAUUUGGGCGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA- .....(....(----((....)))....)....-.--------.....----((..(((((((.((((((......)).))))...)))))))..)).......- ( -13.90) >consensus ____CCUUUUU____GGUUUUUCAUUUUCAUUU_U________AAUCG____UAUUUGGACGCAUUUGCAGCGUUAUGACAAACAAGUGUCCAUUUAUUUGCCA_ ........................................................(((((((.((((((......)).))))...)))))))............ (-11.85 = -11.47 + -0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:32:10 2006