
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,800,368 – 24,800,484 |
| Length | 116 |
| Max. P | 0.893961 |

| Location | 24,800,368 – 24,800,462 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.12 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24800368 94 + 27905053 CUCCUU-------GGCAUCC--UUGAGAAACGGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUUAAA----UUUUUCGGCUAC .....(-------(((....--.(((((((......((((((.....))))))..((((((...(((((((....)))))))...))))))----))))))))))). ( -19.70) >DroVir_CAF1 44431 87 + 1 GUCC-U----------GUCCUGUCCACAAGCUUCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUACCGCAGUCGGCAUAAAUUUAAA------UUUGCGGC--- ...(-(----------((..((((....((((.((....))))))....))))..((((((((.(((.(((....))).)))))))))))------...)))).--- ( -13.30) >DroGri_CAF1 44895 89 + 1 GUCC---------GUCGCCUUGCACACAAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAA------UUUUCGGC--- ....---------...(((.......(((((.((........)).))))).....((((((((.(((((((....)))))))))))))))------.....)))--- ( -22.80) >DroMoj_CAF1 43510 90 + 1 GUCC-U-------GCCGUCCUACACACAAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAA------UUUUCCGC--- ....-.-------.............(((((.((........)).))))).....((((((((.(((((((....)))))))))))))))------........--- ( -18.90) >DroAna_CAF1 10007 100 + 1 CUGCCUGGUAGCUGGUGGCU--GCCACAAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAAA-----UUUUCGGCUAC ..((((((((((.....)))--))))(((((.((........)).)))))....(((((((((.(((((((....))))))))))))))))-----.....)))... ( -34.90) >DroPer_CAF1 11818 97 + 1 CUGC-U-------GGCUGCU--GCCACAUUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAAAUUUUCCUUUCGGCUAC .((.-(-------(((....--)))))).............((((((..((...(((((((((.(((((((....))))))))))))))))...))...)))))).. ( -26.90) >consensus CUCC_U_______GGCGUCC__CCCACAAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAA______UUUUCGGC___ ................................((..((((((.....))))))..((((((((.(((((((....)))))))))))))))............))... (-14.62 = -15.12 + 0.50)



| Location | 24,800,386 – 24,800,484 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.18 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -16.35 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24800386 98 + 27905053 AAACGGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUUAAAUUUUUCGGCUACCGC----UUC-AA-------------GAUGCUGAUGCUCU ....(((((((.((..((((..((.(..((((((...(((((((....)))))))...))))))..).))(((....))----)))-))-------------..)).))))))).. ( -23.70) >DroVir_CAF1 44447 83 + 1 AAGCUUCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUACCGCAGUCGGCAUAAAUUUAAA--UUUGCGGC-------------------------------UGCUGAUGCCGU ..((.((..((((((.....)))))).................(((((((.((..........--..))))))-------------------------------))).)).))... ( -17.70) >DroGri_CAF1 44913 83 + 1 AAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAA--UUUUCGGC-------------------------------UGUUGAUGCUGU .....((((((.(.((((((........((((((((.(((((((....)))))))))))))))--...)))))-------------------------------)).))))))... ( -23.30) >DroWil_CAF1 10081 111 + 1 AAUCUGCGUUAUCGAAUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUUGGCAUAAAUUUAAAA-UUUUAGGCUGCCUU----CUCUGAUGAUGGUGAUGUUGAGGCUGAUGCUCU ..((.(((((((((.............(((((((((.(((((((....))))))))))))))))-..(((((.......----.)))))...))))))))).))(((....))).. ( -24.80) >DroMoj_CAF1 43529 83 + 1 AAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAA--UUUUCCGC-------------------------------UGCUGAUGCUGU .....(((.((((((.....))))))..((((((((.(((((((....)))))))))))))))--.....)))-------------------------------.((....))... ( -20.40) >DroAna_CAF1 10032 101 + 1 AAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAAA-UUUUCGGCUACCGCCGCUUCC-AA-------------GAUGUUGAUGCUCU .....((((((.((..(((...((...(((((((((.(((((((....))))))))))))))))-...))(((....))).....)-))-------------..)).))))))... ( -27.80) >consensus AAUCUGCGUUAUCGAGUUGAUUGAUAUUUUUAAAUUAUGUGCCGCAGUCGGCAUAAAUUUAAA__UUUUCGGC_______________________________UGCUGAUGCUCU .....((((((((((.....)))))...((((((((.(((((((....))))))))))))))).............................................)))))... (-16.35 = -16.72 + 0.36)



| Location | 24,800,386 – 24,800,484 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.18 |
| Mean single sequence MFE | -17.90 |
| Consensus MFE | -11.60 |
| Energy contribution | -11.97 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24800386 98 - 27905053 AGAGCAUCAGCAUC-------------UU-GAA----GCGGUAGCCGAAAAAUUUAAAAUUUAUGCCGACUGCGGCACAUAAUUUAAAAAUAUCAAUCAACUCGAUAACGCCGUUU (((((....)).))-------------).-.((----(((((..........((((((.....(((((....))))).....))))))..((((.........))))..))))))) ( -18.70) >DroVir_CAF1 44447 83 - 1 ACGGCAUCAGCA-------------------------------GCCGCAAA--UUUAAAUUUAUGCCGACUGCGGUACAUAAUUUAAAAAUAUCAAUCAACUCGAUAACGAAGCUU ..(((.......-------------------------------)))((...--((((((((..(((((....)))))...)))))))).............(((....))).)).. ( -16.60) >DroGri_CAF1 44913 83 - 1 ACAGCAUCAACA-------------------------------GCCGAAAA--UUUAAAUUUAUGCCGACUGCGGCACAUAAUUUAAAAAUAUCAAUCAACUCGAUAACGCAGAUU ...((.......-------------------------------........--((((((((..(((((....)))))...))))))))..((((.........))))..))..... ( -14.60) >DroWil_CAF1 10081 111 - 1 AGAGCAUCAGCCUCAACAUCACCAUCAUCAGAG----AAGGCAGCCUAAAA-UUUUAAAUUUAUGCCAACUGCGGCACAUAAUUUAAAAAUAUCAAUCAAUUCGAUAACGCAGAUU .....(((.((((.....((..........)).----.)))).((......-(((((((((..((((......))))...))))))))).((((.........))))..)).))). ( -17.40) >DroMoj_CAF1 43529 83 - 1 ACAGCAUCAGCA-------------------------------GCGGAAAA--UUUAAAUUUAUGCCGACUGCGGCACAUAAUUUAAAAAUAUCAAUCAACUCGAUAACGCAGAUU ...((....)).-------------------------------(((.....--((((((((..(((((....)))))...))))))))..((((.........)))).)))..... ( -18.00) >DroAna_CAF1 10032 101 - 1 AGAGCAUCAACAUC-------------UU-GGAAGCGGCGGUAGCCGAAAA-UUUUAAAUUUAUGCCGACUGCGGCACAUAAUUUAAAAAUAUCAAUCAACUCGAUAACGCAGAUU ...((.........-------------..-.....((((....))))....-(((((((((..(((((....)))))...))))))))).((((.........))))..))..... ( -22.10) >consensus ACAGCAUCAGCA_______________________________GCCGAAAA__UUUAAAUUUAUGCCGACUGCGGCACAUAAUUUAAAAAUAUCAAUCAACUCGAUAACGCAGAUU ...((................................................((((((((..(((((....)))))...))))))))..((((.........))))..))..... (-11.60 = -11.97 + 0.36)

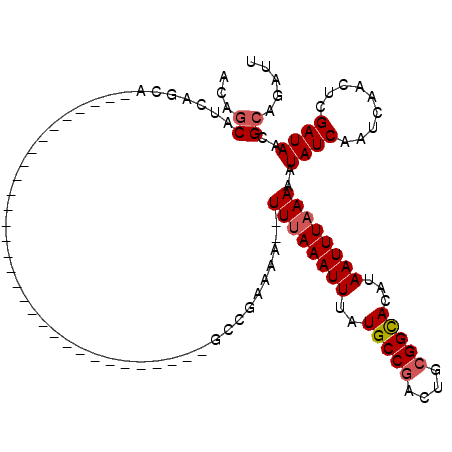
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:32:02 2006