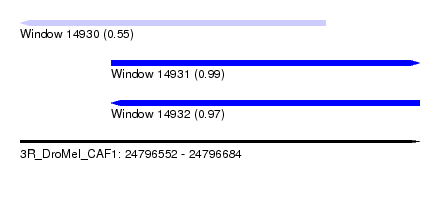
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,796,552 – 24,796,684 |
| Length | 132 |
| Max. P | 0.985194 |

| Location | 24,796,552 – 24,796,653 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.43 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.57 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24796552 101 - 27905053 GGCAAACGCGAUUGCAGCAAAUCAGAAAAUACUCGC--ACAAGUGCGAAUGUGAAUAUUGAUGCCACGGC-----GUA--GGU-------AUGUUUGACUUA---UAAUAUAUGUAUAGA (((....((.......))..(((((...(((.((((--(....))))).))).....)))))))).....-----(((--.((-------(((((.......---.))))))).)))... ( -23.10) >DroPse_CAF1 6884 111 - 1 GGCUGACGCGAUUGCAGCAAAUCAGAAAAUACUCGC--ACAAGUGCGAAUGUGAAUAUUGAUGCCAUGGC-----GUAUAAGUAUUAGGAGUGUAUGAGUGUUGCUUGUGUGUCU--UCU ((..((((((...((((((..(((....((((((((--(....))).......((((((.((((....))-----))...))))))..)))))).))).))))))...)))))).--.)) ( -35.80) >DroWil_CAF1 5861 96 - 1 GGCUG-CGCGAUUGCAGCAAAUCAGAAAAUACUCGCUCACAAGUGCGAAUGUGAAUAUUGAUGGCAAACC-----AAA--AGA-------AUGUUUC---UA---U-AUACAUAU--AUC .((((-((....))))))..............((((........))))(((((.((((.((.((((....-----...--...-------.))))))---.)---)-)).)))))--... ( -19.10) >DroYak_CAF1 7736 98 - 1 GGCAAACGCGAUUGCAGCAAAUCAGAAAAUACUCGC--ACAAGUGCGAAUGUGAAUAUUGAUGCCACGGC-----GUA--GGU-------AUGUAUGGCUU----UAAUAUAUAU--UAU ......(((((((......)))).........((((--(....)))))..))).(((((((.((((..((-----((.--...-------)))).)))).)----))))))....--... ( -23.60) >DroAna_CAF1 5587 104 - 1 GGCAGACGCGAUUGCAGCAAAUCAGAAAAUACUCGC--ACAAGUGCGAAUGUGAAUAUUGAUGCCGUGGCAUCGCAUA--AGU-------CUGUAUGUGCGA---UUAUGUACAU--AAA .(((((((((((.((.((..(((((...(((.((((--(....))))).))).....)))))...)).)))))))...--.))-------))))((((((..---....))))))--... ( -33.30) >DroPer_CAF1 7036 111 - 1 GGCUGACGCGAUUGCAGCAAAUCAGAAAAUACUCGC--ACAAGUGCGAAUGUGAAUAUUGAUGCCAUGGC-----GUAUAAGUAUUAGGAGUGUAUGAGUGUUGCUUGUGUGUCU--UCU ((..((((((...((((((..(((....((((((((--(....))).......((((((.((((....))-----))...))))))..)))))).))).))))))...)))))).--.)) ( -35.80) >consensus GGCAGACGCGAUUGCAGCAAAUCAGAAAAUACUCGC__ACAAGUGCGAAUGUGAAUAUUGAUGCCAUGGC_____GUA__AGU_______AUGUAUGAGUGA___UUAUAUAUAU__AAU (((....((.......))..(((((...(((.((((........)))).))).....))))))))....................................................... (-12.40 = -12.57 + 0.17)



| Location | 24,796,582 – 24,796,684 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 96.14 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.55 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24796582 102 + 27905053 UAC-----GCCGUGGCAUCAAUAUUCACAUUCGCACUUGUGCGAGUAUUUUCUGAUUUGCUGCAAUCGCGUUUGCCAACAACGAACUUGGCUAACAACAACAGCUGC ...-----((.(..(((...........(((((((....)))))))...........)))..)......(((.(((((........))))).))).......))... ( -25.65) >DroSec_CAF1 12852 102 + 1 UAC-----GCCGUGGCAUCAAUAUUCACAUUCGCACUUGUGCGAGUAUUUUCUGAUUUGCUGCAAUCGCGUUUGCCAACAACGAACUUGGCUAACAACAACAGCUGC ...-----((.(..(((...........(((((((....)))))))...........)))..)......(((.(((((........))))).))).......))... ( -25.65) >DroSim_CAF1 12899 102 + 1 UAC-----GCCGUGGCAUCAAUAUUCACAUUCGCACUUGUGCGAGUAUUUUCUGAUUUGCUGCAAUCGCGUUUGCCAACAACGAACUUGGCUAACAACAACAGCUGC ...-----((.(..(((...........(((((((....)))))))...........)))..)......(((.(((((........))))).))).......))... ( -25.65) >DroEre_CAF1 7528 102 + 1 UAC-----GCCGUGGCAUCAAUAUUCACAUUCGCACUUGUGCGAGUAUUCUCUGAUUUGCUGCAAUCGCGUUUGCCAACAACGAACUUGGCUAACAACAACAGCUGC ...-----((.(..(((...........(((((((....)))))))...........)))..)......(((.(((((........))))).))).......))... ( -25.65) >DroYak_CAF1 7763 102 + 1 UAC-----GCCGUGGCAUCAAUAUUCACAUUCGCACUUGUGCGAGUAUUUUCUGAUUUGCUGCAAUCGCGUUUGCCAACAACGAACUUGGCUAACAACAACAGCUGC ...-----((.(..(((...........(((((((....)))))))...........)))..)......(((.(((((........))))).))).......))... ( -25.65) >DroAna_CAF1 5615 107 + 1 UAUGCGAUGCCACGGCAUCAAUAUUCACAUUCGCACUUGUGCGAGUAUUUUCUGAUUUGCUGCAAUCGCGUCUGCCAACAACAAACUUGGCUAACAACAACAGCUUC ...(((((....(((((...........(((((((....)))))))...........)))))..)))))((..(((((........)))))..))............ ( -29.35) >consensus UAC_____GCCGUGGCAUCAAUAUUCACAUUCGCACUUGUGCGAGUAUUUUCUGAUUUGCUGCAAUCGCGUUUGCCAACAACGAACUUGGCUAACAACAACAGCUGC ........((.((((((...........(((((((....)))))))...........))))))......(((.(((((........))))).))).......))... (-24.35 = -24.55 + 0.20)



| Location | 24,796,582 – 24,796,684 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 96.14 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.48 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24796582 102 - 27905053 GCAGCUGUUGUUGUUAGCCAAGUUCGUUGUUGGCAAACGCGAUUGCAGCAAAUCAGAAAAUACUCGCACAAGUGCGAAUGUGAAUAUUGAUGCCACGGC-----GUA ...(((((.(((((..(((((........)))))....))))).)))))..(((((...(((.(((((....))))).))).....)))))((....))-----... ( -29.90) >DroSec_CAF1 12852 102 - 1 GCAGCUGUUGUUGUUAGCCAAGUUCGUUGUUGGCAAACGCGAUUGCAGCAAAUCAGAAAAUACUCGCACAAGUGCGAAUGUGAAUAUUGAUGCCACGGC-----GUA ...(((((.(((((..(((((........)))))....))))).)))))..(((((...(((.(((((....))))).))).....)))))((....))-----... ( -29.90) >DroSim_CAF1 12899 102 - 1 GCAGCUGUUGUUGUUAGCCAAGUUCGUUGUUGGCAAACGCGAUUGCAGCAAAUCAGAAAAUACUCGCACAAGUGCGAAUGUGAAUAUUGAUGCCACGGC-----GUA ...(((((.(((((..(((((........)))))....))))).)))))..(((((...(((.(((((....))))).))).....)))))((....))-----... ( -29.90) >DroEre_CAF1 7528 102 - 1 GCAGCUGUUGUUGUUAGCCAAGUUCGUUGUUGGCAAACGCGAUUGCAGCAAAUCAGAGAAUACUCGCACAAGUGCGAAUGUGAAUAUUGAUGCCACGGC-----GUA ...(((((.(((((..(((((........)))))....))))).)))))..(((((...(((.(((((....))))).))).....)))))((....))-----... ( -29.90) >DroYak_CAF1 7763 102 - 1 GCAGCUGUUGUUGUUAGCCAAGUUCGUUGUUGGCAAACGCGAUUGCAGCAAAUCAGAAAAUACUCGCACAAGUGCGAAUGUGAAUAUUGAUGCCACGGC-----GUA ...(((((.(((((..(((((........)))))....))))).)))))..(((((...(((.(((((....))))).))).....)))))((....))-----... ( -29.90) >DroAna_CAF1 5615 107 - 1 GAAGCUGUUGUUGUUAGCCAAGUUUGUUGUUGGCAGACGCGAUUGCAGCAAAUCAGAAAAUACUCGCACAAGUGCGAAUGUGAAUAUUGAUGCCGUGGCAUCGCAUA ...((.(((((.(((.(((((........))))).)))))))).)).((..........(((.(((((....))))).))).......(((((....)))))))... ( -33.20) >consensus GCAGCUGUUGUUGUUAGCCAAGUUCGUUGUUGGCAAACGCGAUUGCAGCAAAUCAGAAAAUACUCGCACAAGUGCGAAUGUGAAUAUUGAUGCCACGGC_____GUA ...(((((.(((((..(((((........)))))....))))).)))))..(((((...(((.(((((....))))).))).....)))))((....))........ (-28.48 = -28.48 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:31:58 2006