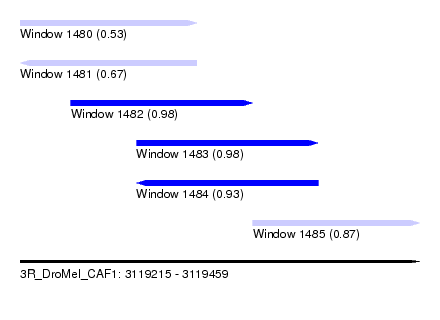
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,119,215 – 3,119,459 |
| Length | 244 |
| Max. P | 0.981943 |

| Location | 3,119,215 – 3,119,323 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.28 |
| Mean single sequence MFE | -40.38 |
| Consensus MFE | -22.85 |
| Energy contribution | -24.72 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
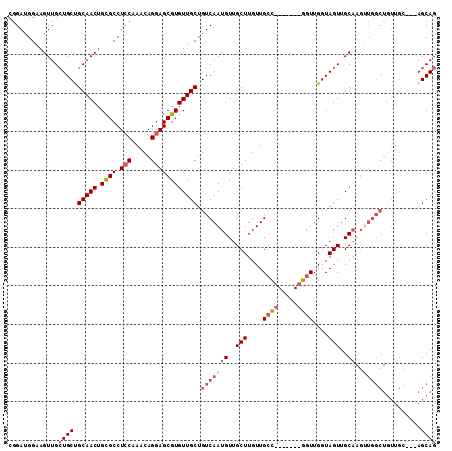
>3R_DroMel_CAF1 3119215 108 + 27905053 CGGAUGGAAGUUGCUGCUGCAACUGCGCCUGCAAACAGGAGCGUGUUGCUGUCAAUGUUGCUUGUUGCCGCUUGGCGGUUGGUAGUUGCAAGUUGGCUGUUGC---AGCAG ...........((((((.((((((((.((((....)))).))).))))).((((((((.(((....(((((...)))))....))).)))..)))))....))---)))). ( -41.70) >DroSec_CAF1 6852 101 + 1 CGGAUGGAAGUUGCUGCCGCAACUGCGCCUCCAAACAGGAGCGUGUUGCUGUCAAUGUUGCUUGUUGCC-------GGUUGGUAGUUGCAAGUUGGCUGUUGC---AGCAG ...........((((((.(((((.((((.(((.....)))))))))))).((((((.((((...(((((-------....)))))..))))))))))....))---)))). ( -39.10) >DroSim_CAF1 12166 101 + 1 CGGAUGGAAGUUGGUGCCGCAACUGCGCCUCCAAACAGGAGCGUGUUGCUGUCAAUGUUGCUUGUUGCC-------GGUUGGUAGUUGCAAGUUGGCUGUUGC---AGCAG ...........((.(((.(((((.((((.(((.....)))))))))))).((((((.((((...(((((-------....)))))..))))))))))....))---).)). ( -34.60) >DroEre_CAF1 6611 87 + 1 CGGAUGGAAGUUGCUGCUGCAACUGCGCCUCCGAACAGGAGCGUGUUGCUGUCAAUGUGGCUUGUUGGC-------GGUUGGUAGUUGC-----------------AGCAG .............(((((((((((((((((((.....)))).))...((((((((((.....)))))))-------)))..))))))))-----------------))))) ( -39.60) >DroYak_CAF1 10532 105 + 1 CGGAGGGAAGUUGCUGCUGCAACAGUGCCUCCAAGCAGGAGCGUGUUGCUGUCAAUGUUGCUUGUUGCAA------UGUUGGUAGUUGCAAGCUGGCUGUUGAAGCAGCAG ...........(((((((.(((((((.(((......)))(((.(((.(((..(((((((((.....))))------)))))..))).))).))).))))))).))))))). ( -46.90) >consensus CGGAUGGAAGUUGCUGCUGCAACUGCGCCUCCAAACAGGAGCGUGUUGCUGUCAAUGUUGCUUGUUGCC_______GGUUGGUAGUUGCAAGUUGGCUGUUGC___AGCAG .............((((.(((((.((((.(((.....)))))))))))).(((((((..(((....((((.....))))....)))..))..)))))..........)))) (-22.85 = -24.72 + 1.87)

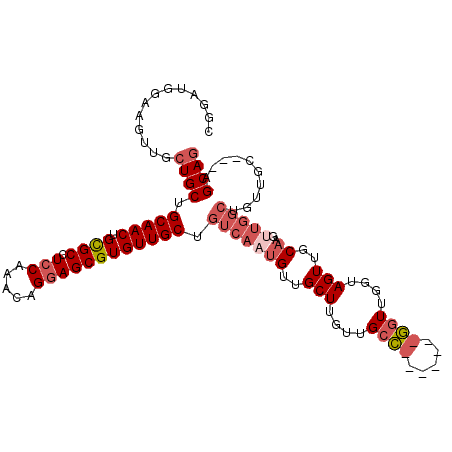
| Location | 3,119,215 – 3,119,323 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.28 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3119215 108 - 27905053 CUGCU---GCAACAGCCAACUUGCAACUACCAACCGCCAAGCGGCAACAAGCAACAUUGACAGCAACACGCUCCUGUUUGCAGGCGCAGUUGCAGCAGCAACUUCCAUCCG (((((---(((((.......((((...........(((....)))..(((......)))...))))...((.((((....)))).)).))))))))))............. ( -36.30) >DroSec_CAF1 6852 101 - 1 CUGCU---GCAACAGCCAACUUGCAACUACCAACC-------GGCAACAAGCAACAUUGACAGCAACACGCUCCUGUUUGGAGGCGCAGUUGCGGCAGCAACUUCCAUCCG (((((---(((((.(((...((((.....((....-------))...(((......)))...)))).....(((.....))))))...))))))))))............. ( -29.20) >DroSim_CAF1 12166 101 - 1 CUGCU---GCAACAGCCAACUUGCAACUACCAACC-------GGCAACAAGCAACAUUGACAGCAACACGCUCCUGUUUGGAGGCGCAGUUGCGGCACCAACUUCCAUCCG .((((---(((((.(((...((((.....((....-------))...(((......)))...)))).....(((.....))))))...))))))))).............. ( -26.80) >DroEre_CAF1 6611 87 - 1 CUGCU-----------------GCAACUACCAACC-------GCCAACAAGCCACAUUGACAGCAACACGCUCCUGUUCGGAGGCGCAGUUGCAGCAGCAACUUCCAUCCG (((((-----------------((((((.......-------((...(((......)))...))....((((((.....))).))).)))))))))))............. ( -27.40) >DroYak_CAF1 10532 105 - 1 CUGCUGCUUCAACAGCCAGCUUGCAACUACCAACA------UUGCAACAAGCAACAUUGACAGCAACACGCUCCUGCUUGGAGGCACUGUUGCAGCAGCAACUUCCCUCCG .(((((((.((((((((...((((((.........------))))))((((((........(((.....)))..))))))..)))..))))).)))))))........... ( -32.60) >consensus CUGCU___GCAACAGCCAACUUGCAACUACCAACC_______GGCAACAAGCAACAUUGACAGCAACACGCUCCUGUUUGGAGGCGCAGUUGCAGCAGCAACUUCCAUCCG (((((..........................................(((......)))...(((((.((((((.....))).)))..))))))))))............. (-16.96 = -17.32 + 0.36)

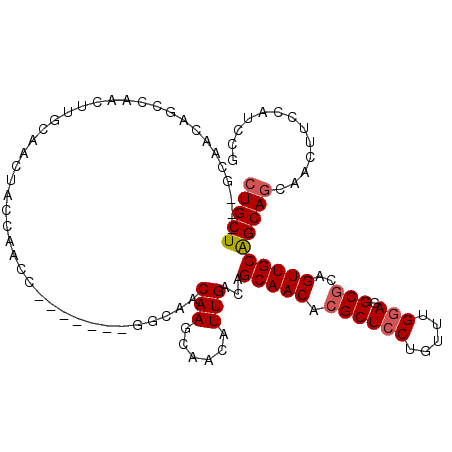
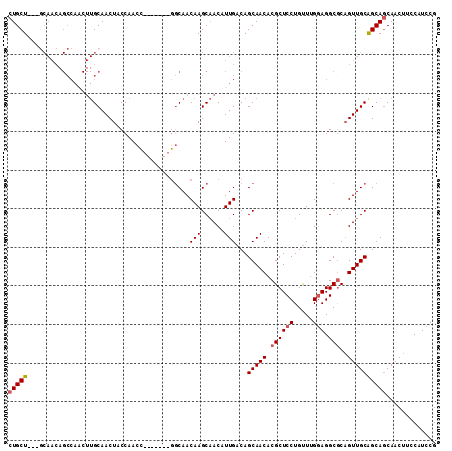
| Location | 3,119,246 – 3,119,357 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -42.12 |
| Consensus MFE | -34.53 |
| Energy contribution | -36.27 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3119246 111 + 27905053 CAAACAGGAGCGUGUUGCUGUCAAUGUUGCUUGUUGCCGCUUGGCGGUUGGUAGUUGCAAGUUGGCUGUUGC---AGCAGCAACAAUUGCGCUACCAGCGAGCCAGCCAGGCUA ..(((((((((((..........))))).))))))((((((.(((.(((((((((.((((....(((((...---.))))).....)))))))))))))..))))))..))).. ( -46.90) >DroSec_CAF1 6883 104 + 1 CAAACAGGAGCGUGUUGCUGUCAAUGUUGCUUGUUGCC-------GGUUGGUAGUUGCAAGUUGGCUGUUGC---AGCAGCAGCAAUUGCGCUACCAGCGAGUCAGCCAGGCUA ........(((.((..((((.(......((.....)).-------.(((((((((.((((....(((((((.---..)))))))..)))))))))))))..).)))))).))). ( -40.50) >DroSim_CAF1 12197 104 + 1 CAAACAGGAGCGUGUUGCUGUCAAUGUUGCUUGUUGCC-------GGUUGGUAGUUGCAAGUUGGCUGUUGC---AGCAGCAGCAAUUGCGCUACCAGCGAGUCAGCCAGGCUA ........(((.((..((((.(......((.....)).-------.(((((((((.((((....(((((((.---..)))))))..)))))))))))))..).)))))).))). ( -40.50) >DroYak_CAF1 10563 103 + 1 CAAGCAGGAGCGUGUUGCUGUCAAUGUUGCUUGUUGCAA------UGUUGGUAGUUGCAAGCUGGCUGUUGAAGCAGCAGCAACAAUC-CGCUACCAGCAA----GCCAGGCUA ..(((..(.((.(((.(((..(((((((((.....))))------)))))..))).))).(((((((((....)))))(((.......-.)))..))))..----)))..))). ( -40.60) >consensus CAAACAGGAGCGUGUUGCUGUCAAUGUUGCUUGUUGCC_______GGUUGGUAGUUGCAAGUUGGCUGUUGC___AGCAGCAACAAUUGCGCUACCAGCGAGUCAGCCAGGCUA ........(((.((..((((.(......((.....)).........(((((((((.((((....(((((((......)))))))..)))))))))))))..).)))))).))). (-34.53 = -36.27 + 1.75)

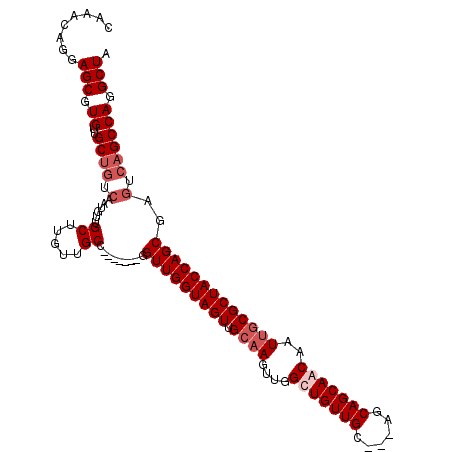
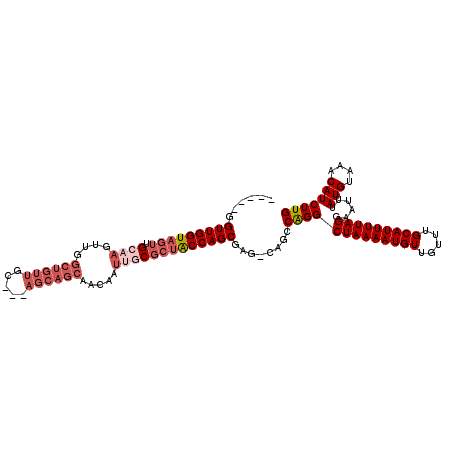
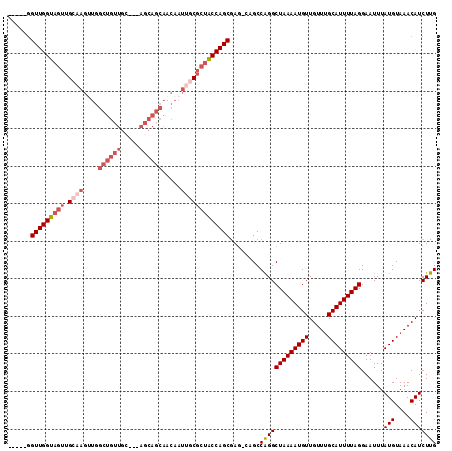
| Location | 3,119,286 – 3,119,397 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -25.60 |
| Energy contribution | -28.08 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3119286 111 + 27905053 UUGGCGGUUGGUAGUUGCAAGUUGGCUGUUGC---AGCAGCAACAAUUGCGCUACCAGCGAGCCAGCCAGGCUAAAAUGUUGUUUGCAUUUUAGGAAUUUAUGUAAACAUCUUG (((((.(((((((((.((((....(((((...---.))))).....)))))))))))))..))))).(((((((((((((.....)))))))))......(((....))))))) ( -41.70) >DroSec_CAF1 6921 106 + 1 -----GGUUGGUAGUUGCAAGUUGGCUGUUGC---AGCAGCAGCAAUUGCGCUACCAGCGAGUCAGCCAGGCUAAAAUGUUGUUUGCAUUUUAGGAAUUUAUGUAAACAUCUUG -----.(((((((((.((((....(((((((.---..)))))))..))))))))))))).((((.....)))).......(((((((((...........)))))))))..... ( -37.80) >DroSim_CAF1 12235 106 + 1 -----GGUUGGUAGUUGCAAGUUGGCUGUUGC---AGCAGCAGCAAUUGCGCUACCAGCGAGUCAGCCAGGCUAAAAUGUUGUUUGCAUUUUAGGAAUUUAUGUAAACAUCUUG -----.(((((((((.((((....(((((((.---..)))))))..))))))))))))).((((.....)))).......(((((((((...........)))))))))..... ( -37.80) >DroEre_CAF1 6680 85 + 1 -----GGUUGGUAGUUGC-----------------AGCAGCAACCAUU-C--CGCCAGCGA----GCCGGGCUAAAAUGUUGUUUGCAUUUUAGGAAUUUAUGUAAACAUCUUG -----(((((.(.((...-----------------.))).)))))(((-(--((((.(...----.)..)))((((((((.....)))))))))))))................ ( -20.60) >DroYak_CAF1 10602 104 + 1 -----UGUUGGUAGUUGCAAGCUGGCUGUUGAAGCAGCAGCAACAAUC-CGCUACCAGCAA----GCCAGGCUAAAAUGUUGUUAGCAUUUUAGGAAUUUAUGUAAACAUCUUG -----((((((((((((...((((.((((....))))))))..))...-.)))))))))).----..(((((((((((((.....)))))))))......(((....))))))) ( -31.70) >consensus _____GGUUGGUAGUUGCAAGUUGGCUGUUGC___AGCAGCAACAAUUGCGCUACCAGCGAG_CAGCCAGGCUAAAAUGUUGUUUGCAUUUUAGGAAUUUAUGUAAACAUCUUG ......(((((((((.((((....((((((.....)))))).....)))))))))))))........(((((((((((((.....)))))))))......(((....))))))) (-25.60 = -28.08 + 2.48)

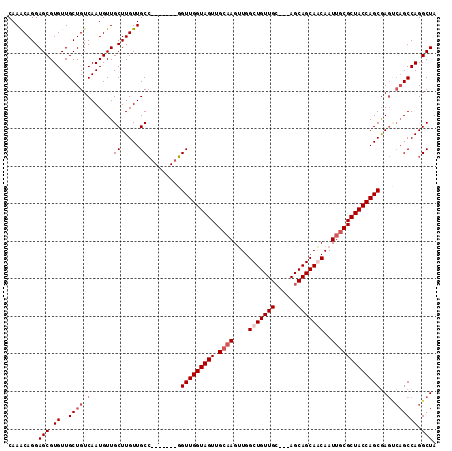
| Location | 3,119,286 – 3,119,397 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -17.32 |
| Energy contribution | -18.70 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
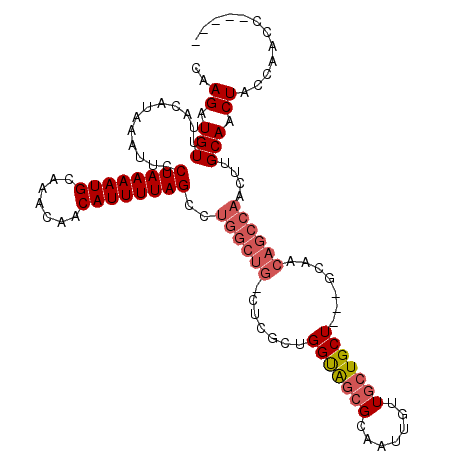
>3R_DroMel_CAF1 3119286 111 - 27905053 CAAGAUGUUUACAUAAAUUCCUAAAAUGCAAACAACAUUUUAGCCUGGCUGGCUCGCUGGUAGCGCAAUUGUUGCUGCU---GCAACAGCCAACUUGCAACUACCAACCGCCAA ....(((....)))......((((((((.......))))))))..((((.((.....((((((.(((((((((((....---))))))).....))))..)))))).)))))). ( -29.50) >DroSec_CAF1 6921 106 - 1 CAAGAUGUUUACAUAAAUUCCUAAAAUGCAAACAACAUUUUAGCCUGGCUGACUCGCUGGUAGCGCAAUUGCUGCUGCU---GCAACAGCCAACUUGCAACUACCAACC----- ((((................((((((((.......))))))))..((((((....((.((((((((....)).))))))---))..)))))).))))............----- ( -28.40) >DroSim_CAF1 12235 106 - 1 CAAGAUGUUUACAUAAAUUCCUAAAAUGCAAACAACAUUUUAGCCUGGCUGACUCGCUGGUAGCGCAAUUGCUGCUGCU---GCAACAGCCAACUUGCAACUACCAACC----- ((((................((((((((.......))))))))..((((((....((.((((((((....)).))))))---))..)))))).))))............----- ( -28.40) >DroEre_CAF1 6680 85 - 1 CAAGAUGUUUACAUAAAUUCCUAAAAUGCAAACAACAUUUUAGCCCGGC----UCGCUGGCG--G-AAUGGUUGCUGCU-----------------GCAACUACCAACC----- ....(((....)))..((((((((((((.......)))))))(((.((.----...))))))--)-)))((((((....-----------------)))))).......----- ( -20.60) >DroYak_CAF1 10602 104 - 1 CAAGAUGUUUACAUAAAUUCCUAAAAUGCUAACAACAUUUUAGCCUGGC----UUGCUGGUAGCG-GAUUGUUGCUGCUGCUUCAACAGCCAGCUUGCAACUACCAACA----- ((((................((((((((.......)))))))).(((((----(....(((((((-(.......)))))))).....))))))))))............----- ( -27.50) >consensus CAAGAUGUUUACAUAAAUUCCUAAAAUGCAAACAACAUUUUAGCCUGGCUG_CUCGCUGGUAGCGCAAUUGUUGCUGCU___GCAACAGCCAACUUGCAACUACCAACC_____ ..((.(((............((((((((.......))))))))..((((((.......(((((((.......))))))).......))))))....))).))............ (-17.32 = -18.70 + 1.38)

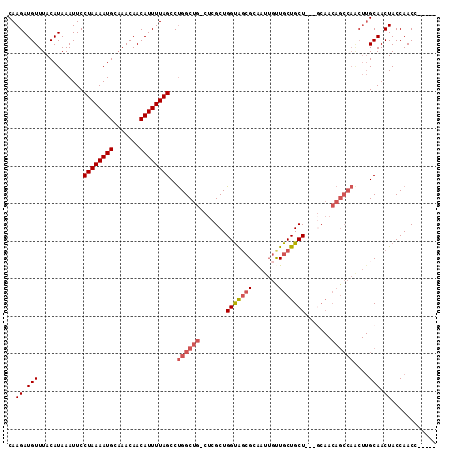
| Location | 3,119,357 – 3,119,459 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.90 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
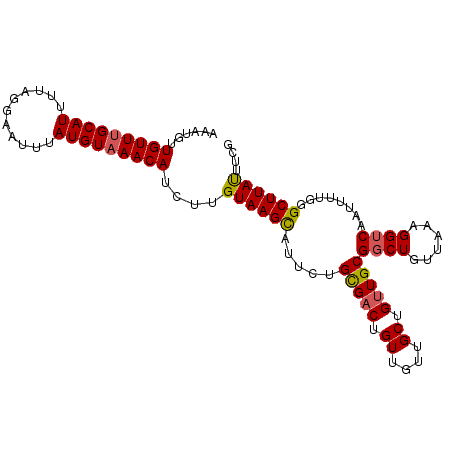
>3R_DroMel_CAF1 3119357 102 + 27905053 AAAUGUUGUUUGCAUUUUAGGAAUUUAUGUAAACAUCUUGUAAGCAUUCUGUGACUGUUGUUGCUGUUGCGGCUGUUAAAGGUCAAUUUUGGGCUUAUUUCG ......(((((((((...........)))))))))....((((((......(((((...(((((....))))).......))))).......)))))).... ( -24.02) >DroSec_CAF1 6987 102 + 1 AAAUGUUGUUUGCAUUUUAGGAAUUUAUGUAAACAUCUUGUAAGCAUUCUGCGACUGUUGUUGCUGUUGCGGCUGUUAAAGGACAAUUUUGGGCUUAUUUCU ......(((((((((...........)))))))))....((((((...(((((((.((....)).))))))).((((....)))).......)))))).... ( -23.90) >DroSim_CAF1 12301 102 + 1 AAAUGUUGUUUGCAUUUUAGGAAUUUAUGUAAACAUCUUGUAAGCAUUCUGCGACUGUUGUUGCUGUUGCGGCUGUUAAAGGUCAAUUUUGGGCUUAUUUCG ......(((((((((...........)))))))))....((((((.....(((((.((....)).)))))((((......))))........)))))).... ( -24.10) >DroEre_CAF1 6725 102 + 1 AAAUGUUGUUUGCAUUUUAGGAAUUUAUGUAAACAUCUUGUAAGCAUUCGGCGACUGUUGUUGCUGCUGCGGCUGCUAAAGGUCAAUUUUCAGCUUACUUCG ......(((((((((...........)))))))))....((((((...(((((((....)))))))....((((......))))........)))))).... ( -27.90) >DroYak_CAF1 10666 102 + 1 AAAUGUUGUUAGCAUUUUAGGAAUUUAUGUAAACAUCUUGUAAGUAUUCUGCGACUGUUGUUGCUGUUGCGGCUGCUAAAGGUCAGUUUUCGGCUUAUUUCG ((((((.....))))))..(((((...((.(((((((((...((((..(((((((.((....)).))))))).)))).)))))..)))).))....))))). ( -22.10) >consensus AAAUGUUGUUUGCAUUUUAGGAAUUUAUGUAAACAUCUUGUAAGCAUUCUGCGACUGUUGUUGCUGUUGCGGCUGUUAAAGGUCAAUUUUGGGCUUAUUUCG ......(((((((((...........)))))))))....((((((.....(((((.((....)).)))))((((......))))........)))))).... (-21.66 = -21.78 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:51 2006