| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,788,413 – 24,788,645 |
| Length | 232 |
| Max. P | 0.749247 |

| Location | 24,788,413 – 24,788,528 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -29.57 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
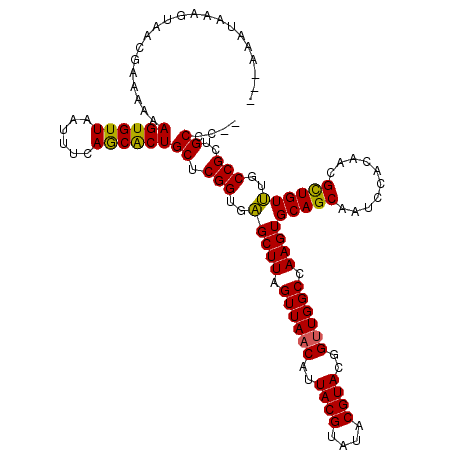
>3R_DroMel_CAF1 24788413 115 + 27905053 CACCCA-----CCUCAUUUCCCUGGGCUGUCUGUCUUGGCAUUGCCAUUUUUACUUCUCAUUGCUUUUGUUUGCCGUGUAAUUAAAAUAAAGGCGUGGAGUAAUUGCCGAGGUCAGCCGA ......-----.............(((((....((((((((.........(((((((....(((((((((((............))))))))))).))))))).)))))))).))))).. ( -33.80) >DroEre_CAF1 28597 112 + 1 CACCCA-----GCUCAUUUCCCUGGGCUGUCUGUCUUGGCAUUGCCAUUUCUACUUCUCGUUGCUUUUGUUUGCCGUGUAAUUAAAAUAAAGGCGUGGAGUAAUUGCCGAGGUCAGU--- ....((-----(((((......))))))).(((((((((((..........((((((....(((((((((((............))))))))))).))))))..)))))))).))).--- ( -35.40) >DroYak_CAF1 20647 117 + 1 CACCCAACCCACCUCAUUUUCCGGGGCUGUCUGUCUUGGCAUUGCCAUUUUUACUUCUCAUUGCUUUUGUUUGCCGUGUAAUUAAAAUAAAGGCGUGGAGUAAUUGCCGAGGUCAGU--- .........((((((.......)))).)).(((((((((((.........(((((((....(((((((((((............))))))))))).))))))).)))))))).))).--- ( -32.00) >consensus CACCCA_____CCUCAUUUCCCUGGGCUGUCUGUCUUGGCAUUGCCAUUUUUACUUCUCAUUGCUUUUGUUUGCCGUGUAAUUAAAAUAAAGGCGUGGAGUAAUUGCCGAGGUCAGU___ (((((..................))).)).(((((((((((.........(((((((....(((((((((((............))))))))))).))))))).)))))))).))).... (-29.57 = -29.90 + 0.33)


| Location | 24,788,528 – 24,788,645 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -30.10 |
| Energy contribution | -29.33 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24788528 117 - 27905053 ---AAAUAAAGUAACGAAAAAAGUGUUAAUUUCAACGCUGCUCGGUGAGCUUAGUUAACAUUACGUAUACGUACGGUUGGCCAAGUGCAGCAAUCCAUAACGCUGUUUGCCGCUGCCGCC ---...........((.....((((((......))))))((.(((..(((((.((((((..((((....))))..)))))).))))(((((..........))))))..)))..)))).. ( -30.20) >DroEre_CAF1 28709 115 - 1 ---AAAUAAAGUAACGAAAAAAGUGUUAAUUUCAGCACUGCUCGGUGCGCUUAGUUAACAUUACGUAUACGUACCGUUGGCCAAGUGCAGCAAUCCACAACGCUGUGUGCCGCUGCCC-- ---.......(((.(((....((((((......))))))..))).)))((((.((((((..((((....))))..)))))).))))(((((....((((....))))....)))))..-- ( -33.10) >DroYak_CAF1 20764 118 - 1 AAAAAAUAAAGUAACGAAAAAAGUGUUAAUUUCAGCACUGCUCGGUGAGCUUAGUUAACAUUACGUAUACGUACGGGUGGCCAAGUGCAGCAAUCCACAACGUUGUUUGCCGCUGCCC-- ..........((((((.....((((((((((....((((....)))).....)))))))))).))).)))....(((..((.....(((((..........))))).....))..)))-- ( -28.90) >consensus ___AAAUAAAGUAACGAAAAAAGUGUUAAUUUCAGCACUGCUCGGUGAGCUUAGUUAACAUUACGUAUACGUACGGUUGGCCAAGUGCAGCAAUCCACAACGCUGUUUGCCGCUGCCC__ .....................((((((......))))))((.(((..(((((.((((((..((((....))))..)))))).))))(((((..........))))))..)))..)).... (-30.10 = -29.33 + -0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:31:46 2006