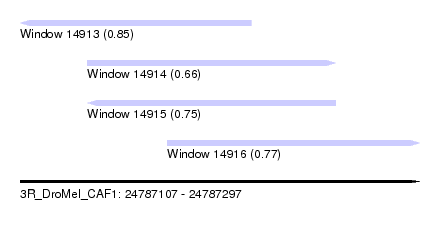
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,787,107 – 24,787,297 |
| Length | 190 |
| Max. P | 0.849829 |

| Location | 24,787,107 – 24,787,217 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.29 |
| Mean single sequence MFE | -17.64 |
| Consensus MFE | -15.47 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24787107 110 - 27905053 AACACAUUCCAUAUCAUUACUGGAGAAGGCCACAAAAAUUUUCUCAAUCCUUAAAUGUGGCAUGCCCCCUCCUAACUC----------ACACACACACCAACAUACAAGGCAGUAACAUC ................(((((((((((((.........)))))))...((((..((((((..((..............----------.....))..)).))))..)))))))))).... ( -15.51) >DroEre_CAF1 27270 120 - 1 AACACAUUCCAUAUCAUUACUGGAGGAGGCCACAAAAAUUUCCUCAAUCCUUAAAUGUGGCAUGCCCCCUCCUAACUCACGCUCACACACACACACACCAGCACACAAGGCAGUAACAUC ................((((((((((..((((((.....................))))))......)))))............................((.......))))))).... ( -18.70) >DroYak_CAF1 19214 116 - 1 AACACAUUCCAUAUCAUUACUGGAGGAGGCCACAAAAAUUUCCUCAAUCCUUAAAUGUGGCAUGCCCCCUCCUAACUUACU----CACACACACACACCAGCACACAAGGCAGUAACAUC ................((((((((((..((((((.....................))))))......))))).........----...............((.......))))))).... ( -18.70) >consensus AACACAUUCCAUAUCAUUACUGGAGGAGGCCACAAAAAUUUCCUCAAUCCUUAAAUGUGGCAUGCCCCCUCCUAACUCAC_____CACACACACACACCAGCACACAAGGCAGUAACAUC ................((((((((((..((((((.....................))))))......))))).........................((.........)).))))).... (-15.47 = -15.80 + 0.33)


| Location | 24,787,139 – 24,787,257 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -30.01 |
| Energy contribution | -29.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24787139 118 + 27905053 --GAGUUAGGAGGGGGCAUGCCACAUUUAAGGAUUGAGAAAAUUUUUGUGGCCUUCUCCAGUAAUGAUAUGGAAUGUGUUGGAAUUUCAAGAAGUUACUCCAGAUUUCUUUAUUUAUUUA --(((((.((((((((...((((((....((((((.....))))))))))))..))))).(((((....((((((........))))))....)))))))).)))))............. ( -27.90) >DroEre_CAF1 27310 120 + 1 GUGAGUUAGGAGGGGGCAUGCCACAUUUAAGGAUUGAGGAAAUUUUUGUGGCCUCCUCCAGUAAUGAUAUGGAAUGUGUUGGAAUUUCAAGAAGUUACUCCAGAUUUCUUUAUUUAUUUA ((((((.(((((((((...((((((....((((((.....))))))))))))..)))))((((((....((((((........))))))....)))))).......)))).))))))... ( -30.90) >DroYak_CAF1 19250 120 + 1 GUAAGUUAGGAGGGGGCAUGCCACAUUUAAGGAUUGAGGAAAUUUUUGUGGCCUCCUCCAGUAAUGAUAUGGAAUGUGUUGGAAUUUCAAGAAGUUACUCCAGAUUUCUUUAUUUAUUUA ((((((.(((((((((...((((((....((((((.....))))))))))))..)))))((((((....((((((........))))))....)))))).......)))).))))))... ( -30.80) >consensus GUGAGUUAGGAGGGGGCAUGCCACAUUUAAGGAUUGAGGAAAUUUUUGUGGCCUCCUCCAGUAAUGAUAUGGAAUGUGUUGGAAUUUCAAGAAGUUACUCCAGAUUUCUUUAUUUAUUUA ..(((((.((((((((...((((((....((((((.....))))))))))))..))))).(((((....((((((........))))))....)))))))).)))))............. (-30.01 = -29.57 + -0.44)

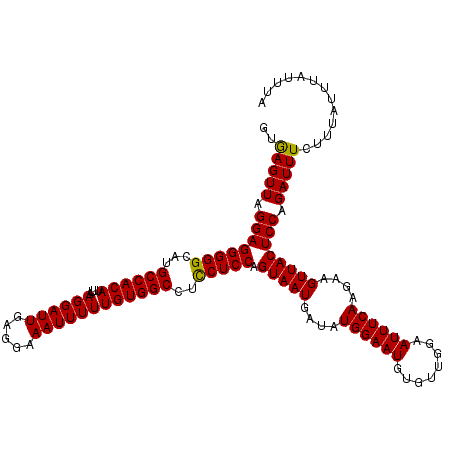
| Location | 24,787,139 – 24,787,257 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -18.59 |
| Energy contribution | -18.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24787139 118 - 27905053 UAAAUAAAUAAAGAAAUCUGGAGUAACUUCUUGAAAUUCCAACACAUUCCAUAUCAUUACUGGAGAAGGCCACAAAAAUUUUCUCAAUCCUUAAAUGUGGCAUGCCCCCUCCUAACUC-- ...................((((...(((((((......))).....((((((....)).))))))))((((((.....................)))))).......))))......-- ( -18.70) >DroEre_CAF1 27310 120 - 1 UAAAUAAAUAAAGAAAUCUGGAGUAACUUCUUGAAAUUCCAACACAUUCCAUAUCAUUACUGGAGGAGGCCACAAAAAUUUCCUCAAUCCUUAAAUGUGGCAUGCCCCCUCCUAACUCAC ............((....((((((..(.....)..))))))....................(((((..((((((.....................))))))......)))))....)).. ( -19.80) >DroYak_CAF1 19250 120 - 1 UAAAUAAAUAAAGAAAUCUGGAGUAACUUCUUGAAAUUCCAACACAUUCCAUAUCAUUACUGGAGGAGGCCACAAAAAUUUCCUCAAUCCUUAAAUGUGGCAUGCCCCCUCCUAACUUAC ..........(((.....((((((..(.....)..))))))....................(((((..((((((.....................))))))......)))))...))).. ( -20.20) >consensus UAAAUAAAUAAAGAAAUCUGGAGUAACUUCUUGAAAUUCCAACACAUUCCAUAUCAUUACUGGAGGAGGCCACAAAAAUUUCCUCAAUCCUUAAAUGUGGCAUGCCCCCUCCUAACUCAC ...................((((...(((((((......))).....((((((....)).))))))))((((((.....................)))))).......))))........ (-18.59 = -18.37 + -0.22)


| Location | 24,787,177 – 24,787,297 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -30.90 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24787177 120 + 27905053 AAUUUUUGUGGCCUUCUCCAGUAAUGAUAUGGAAUGUGUUGGAAUUUCAAGAAGUUACUCCAGAUUUCUUUAUUUAUUUAAAUGAGCAGCCAGCGCGAAUGCCACAAAGUCAGGCCCGUA ...(((((((((.(((((((.........))))..(((((((...((((.((((((......))))))((((......))))))))...)))))))))).)))))))))........... ( -33.50) >DroEre_CAF1 27350 120 + 1 AAUUUUUGUGGCCUCCUCCAGUAAUGAUAUGGAAUGUGUUGGAAUUUCAAGAAGUUACUCCAGAUUUCUUUAUUUAUUUAAAUGAGCAGCCAGCGCGAAUGCCACAAAGUCAGGCCCGUA ...(((((((((....((((.........)))).((((((((...((((.((((((......))))))((((......))))))))...))))))))...)))))))))........... ( -30.90) >DroYak_CAF1 19290 120 + 1 AAUUUUUGUGGCCUCCUCCAGUAAUGAUAUGGAAUGUGUUGGAAUUUCAAGAAGUUACUCCAGAUUUCUUUAUUUAUUUAAAUGAGCAGCCAGCGCGAAUGCCACAAAGUCAGGCCCGUA ...(((((((((....((((.........)))).((((((((...((((.((((((......))))))((((......))))))))...))))))))...)))))))))........... ( -30.90) >consensus AAUUUUUGUGGCCUCCUCCAGUAAUGAUAUGGAAUGUGUUGGAAUUUCAAGAAGUUACUCCAGAUUUCUUUAUUUAUUUAAAUGAGCAGCCAGCGCGAAUGCCACAAAGUCAGGCCCGUA ...(((((((((....((((.........)))).((((((((...((((.((((((......))))))((((......))))))))...))))))))...)))))))))........... (-30.90 = -30.90 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:31:44 2006