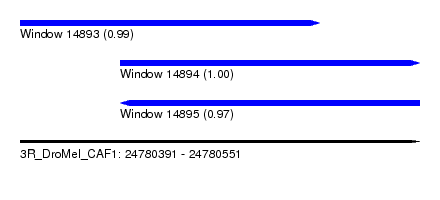
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,780,391 – 24,780,551 |
| Length | 160 |
| Max. P | 0.999536 |

| Location | 24,780,391 – 24,780,511 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -27.54 |
| Energy contribution | -27.30 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24780391 120 + 27905053 CUACAUUCCCUUCUCUGUCUAUCUAUCACUAUCCAUCUCUUUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCAGUUGUACCAGCAAAGGCCAAGCUGUGGCAA ...........................................(((....)))(((..((..((...(((((..(((((((..((....))..)))))))..)))))..)).))..))). ( -27.30) >DroSec_CAF1 11468 118 + 1 CUAG--CUCCCUCUCUGUCUAUCUAUCACUAUCCAUCUCGUUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCAGUUGUACCAGCAAAGGCCAAGCUGUGGCAA ....--.....................................(((....)))(((..((..((...(((((..(((((((..((....))..)))))))..)))))..)).))..))). ( -27.30) >DroSim_CAF1 6836 118 + 1 CUAG--CCCGCUCUCUGUCUAUCUAUCACUAUCCAUCUCUUUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCAGUUGUACCAGCAAAGGCCAAGCUGUGGCAA ...(--((((((...............................(((....)))..............(((((..(((((((..((....))..)))))))..)))))..))).).))).. ( -28.70) >DroEre_CAF1 20489 118 + 1 CUAG--CUCCCUCUCUGUCUAUCUAUCACUAUCCAUCUCUUUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCGGUUGUACCAGCAAAGGCCAAGCUGUGGCAA ....--.....................................(((....)))(((..((..((...(((((..(((((((..((....))..)))))))..)))))..)).))..))). ( -27.30) >DroYak_CAF1 12075 118 + 1 CUAG--CUACCUCUCUGUCUAUCUAUCACUAUCCAUCUCUUUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCGGUUGUACCAGCAAAGGCCAAGCUGUGGCAA ...(--((((..((.((..........................(((....)))..............(((((..(((((((..((....))..)))))))..)))))))))..))))).. ( -28.20) >consensus CUAG__CUCCCUCUCUGUCUAUCUAUCACUAUCCAUCUCUUUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCAGUUGUACCAGCAAAGGCCAAGCUGUGGCAA ...........................................(((....)))(((..((..((...(((((..(((((((..((....))..)))))))..)))))..)).))..))). (-27.54 = -27.30 + -0.24)


| Location | 24,780,431 – 24,780,551 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -38.54 |
| Energy contribution | -38.30 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24780431 120 + 27905053 UUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCAGUUGUACCAGCAAAGGCCAAGCUGUGGCAAAAUUAAAUUACUCAUACGCUCAGUUGGCCAGGGCUCUAUA ...((((((..((((..((........(((((..(((((((..((....))..)))))))..)))))...((....))...................)).))))..))..))))...... ( -38.30) >DroSec_CAF1 11506 120 + 1 UUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCAGUUGUACCAGCAAAGGCCAAGCUGUGGCAAAAUUAAAUUACUCAUACGCUCAGUUGGCCAGGGCUCUAUA ...((((((..((((..((........(((((..(((((((..((....))..)))))))..)))))...((....))...................)).))))..))..))))...... ( -38.30) >DroSim_CAF1 6874 120 + 1 UUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCAGUUGUACCAGCAAAGGCCAAGCUGUGGCAAAAUUAAAUUACUCAUACGCUCAGUUGGCCAGGGCUCUAUA ...((((((..((((..((........(((((..(((((((..((....))..)))))))..)))))...((....))...................)).))))..))..))))...... ( -38.30) >DroEre_CAF1 20527 120 + 1 UUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCGGUUGUACCAGCAAAGGCCAAGCUGUGGCAAAAUUAAAUUACUCAUACGCUCAGUUGGCCAGGGCUCCAUA ...((((((..((((..((........(((((..(((((((..((....))..)))))))..)))))...((....))...................)).))))..))..))))...... ( -38.30) >DroYak_CAF1 12113 120 + 1 UUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCGGUUGUACCAGCAAAGGCCAAGCUGUGGCAAAAUUAAAUUACUCAUACGCUCAGUUGGCCAGGGCUCCAUA ...((((((..((((..((........(((((..(((((((..((....))..)))))))..)))))...((....))...................)).))))..))..))))...... ( -38.30) >consensus UUCGCCUGCUGGCUGUUGCAAUCUCUAGCUUUCUCUGGUAUUCCUGACCAGUUGUACCAGCAAAGGCCAAGCUGUGGCAAAAUUAAAUUACUCAUACGCUCAGUUGGCCAGGGCUCUAUA ...((((((..((((..((........(((((..(((((((..((....))..)))))))..)))))...((....))...................)).))))..))..))))...... (-38.54 = -38.30 + -0.24)


| Location | 24,780,431 – 24,780,551 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -36.52 |
| Energy contribution | -36.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24780431 120 - 27905053 UAUAGAGCCCUGGCCAACUGAGCGUAUGAGUAAUUUAAUUUUGCCACAGCUUGGCCUUUGCUGGUACAACUGGUCAGGAAUACCAGAGAAAGCUAGAGAUUGCAACAGCCAGCAGGCGAA ......(((..(((((((((.(.(((.((((......)))))))).))).))))))..(((((((....(((((.......))))).....((........))....))))))))))... ( -38.60) >DroSec_CAF1 11506 120 - 1 UAUAGAGCCCUGGCCAACUGAGCGUAUGAGUAAUUUAAUUUUGCCACAGCUUGGCCUUUGCUGGUACAACUGGUCAGGAAUACCAGAGAAAGCUAGAGAUUGCAACAGCCAGCAGGCGAA ......(((..(((((((((.(.(((.((((......)))))))).))).))))))..(((((((....(((((.......))))).....((........))....))))))))))... ( -38.60) >DroSim_CAF1 6874 120 - 1 UAUAGAGCCCUGGCCAACUGAGCGUAUGAGUAAUUUAAUUUUGCCACAGCUUGGCCUUUGCUGGUACAACUGGUCAGGAAUACCAGAGAAAGCUAGAGAUUGCAACAGCCAGCAGGCGAA ......(((..(((((((((.(.(((.((((......)))))))).))).))))))..(((((((....(((((.......))))).....((........))....))))))))))... ( -38.60) >DroEre_CAF1 20527 120 - 1 UAUGGAGCCCUGGCCAACUGAGCGUAUGAGUAAUUUAAUUUUGCCACAGCUUGGCCUUUGCUGGUACAACCGGUCAGGAAUACCAGAGAAAGCUAGAGAUUGCAACAGCCAGCAGGCGAA ......(((..(((((((((.(.(((.((((......)))))))).))).))))))..(((((((......(((.......))).......((........))....))))))))))... ( -34.80) >DroYak_CAF1 12113 120 - 1 UAUGGAGCCCUGGCCAACUGAGCGUAUGAGUAAUUUAAUUUUGCCACAGCUUGGCCUUUGCUGGUACAACCGGUCAGGAAUACCAGAGAAAGCUAGAGAUUGCAACAGCCAGCAGGCGAA ......(((..(((((((((.(.(((.((((......)))))))).))).))))))..(((((((......(((.......))).......((........))....))))))))))... ( -34.80) >consensus UAUAGAGCCCUGGCCAACUGAGCGUAUGAGUAAUUUAAUUUUGCCACAGCUUGGCCUUUGCUGGUACAACUGGUCAGGAAUACCAGAGAAAGCUAGAGAUUGCAACAGCCAGCAGGCGAA ......(((..(((((((((.(.(((.((((......)))))))).))).))))))..(((((((....(((((.......))))).....((........))....))))))))))... (-36.52 = -36.92 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:31:26 2006