
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,779,653 – 24,779,790 |
| Length | 137 |
| Max. P | 0.695782 |

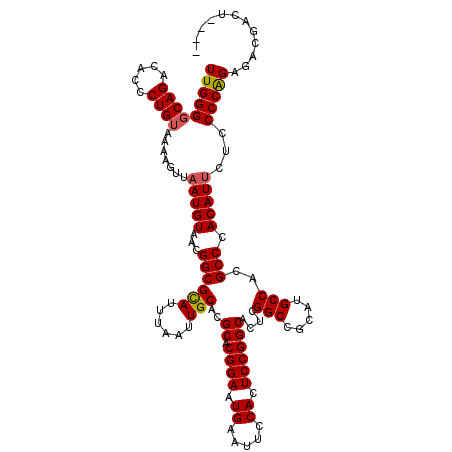
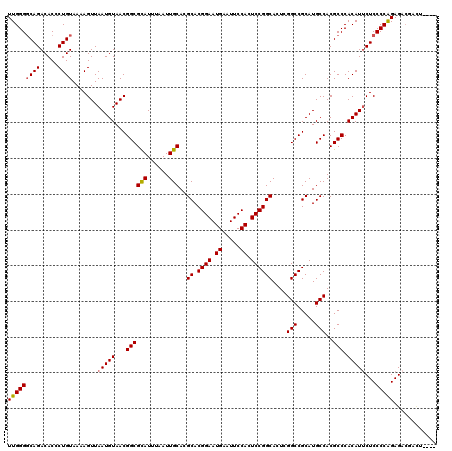
| Location | 24,779,653 – 24,779,766 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -30.82 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24779653 113 + 27905053 UUGGGCCAGACACCCUGUAAAAGUUAAUGUAACGGCGCAUUUAAUUGCACGCACGGAAUGAAUUCCACUCCGGCACUCGGCCGCAUGCCACGCCCACAUUCUCCCCGGAG---ACU---- .((((((((.....)))................((((((......)))..((..(((.((.....)).)))(((.....)))))..)))..)))))...((((....)))---)..---- ( -34.50) >DroSec_CAF1 10735 116 + 1 UUGGGGCAGACACCCUGUAAAAGUUAAUGUAACGGCGCAUUUAAUUGCACGCACGGAAUGAAUUCCACUCCGGCACUCGGCCGCAUGCCACGCCCACAUUCUCCCCAGAGACGACU---- (((((((((.....))))....(..(((((...((((((......)))..((.((((.((.....)).))))))....(((.....)))..))).)))))..))))))........---- ( -33.00) >DroSim_CAF1 6112 116 + 1 UUGGGGCAGACACCCUGUAAAAGUUAAUGUAACGGCGCAUUUAAUUGCACGCACGGAAUGAAUUCCACUCCGGCACUCGGCCGCAUGCCACGCCCACAUUCUCCCCAGAGACGACU---- (((((((((.....))))....(..(((((...((((((......)))..((.((((.((.....)).))))))....(((.....)))..))).)))))..))))))........---- ( -33.00) >DroEre_CAF1 19730 113 + 1 UUGGGGCAGACACCCUGUAAAAGUUAAUGUAACGGCGCAUUUAAUUGCACGCACGGAAUGAAUUCCACUCCGGCACUCGGCCGCAUGCCACGCCCACAUGCUCCCCAGAGACU------- ((((((.((.((...(((....((((...))))((((((......)))..((.((((.((.....)).))))))....(((.....)))..))).))))))))))))).....------- ( -32.60) >DroYak_CAF1 11321 120 + 1 UUGGGGCAGACACCCUGUAAAAGUUAAUGUAACGGCGUAUUUAAUUGCACGCACGGAAUGAAUUCCACUCCGGCACUCGGCCGCAUGCCACGCCCACAUUCUCCCCAGAGACGGUAUUUU (((((((((.....))))....(..(((((...(((((............((.((((.((.....)).))))))....(((.....)))))))).)))))..))))))............ ( -33.30) >consensus UUGGGGCAGACACCCUGUAAAAGUUAAUGUAACGGCGCAUUUAAUUGCACGCACGGAAUGAAUUCCACUCCGGCACUCGGCCGCAUGCCACGCCCACAUUCUCCCCAGAGACGACU____ (((((((((.....)))).......(((((...((((((......)))..((.((((.((.....)).))))))....(((.....)))..))).)))))...)))))............ (-30.82 = -30.90 + 0.08)

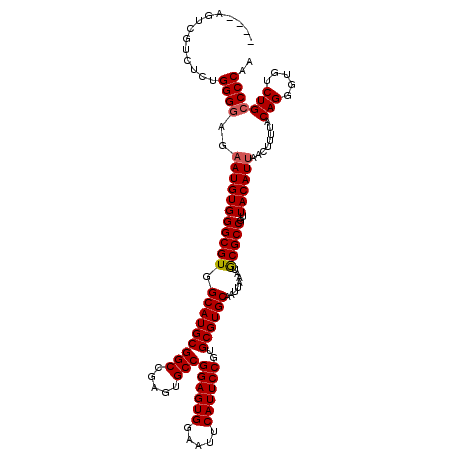
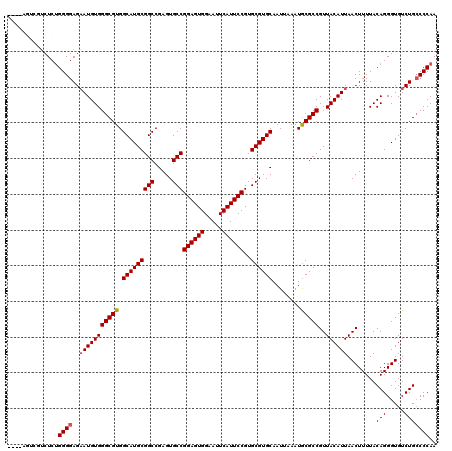
| Location | 24,779,653 – 24,779,766 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -41.48 |
| Consensus MFE | -38.82 |
| Energy contribution | -39.06 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24779653 113 - 27905053 ----AGU---CUCCGGGGAGAAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAAAUGCGCCGUUACAUUAACUUUUACAGGGUGUCUGGCCCAA ----..(---(((....))))...(((((..(((((.(....(((((((((((((.....))))))).))(((((......)))))..........)))).....).)))))..))))). ( -41.40) >DroSec_CAF1 10735 116 - 1 ----AGUCGUCUCUGGGGAGAAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAAAUGCGCCGUUACAUUAACUUUUACAGGGUGUCUGCCCCAA ----.........((((((((.(((((((((.(((((((((.....)))((((((.....))))))..))))))........)))))((((...))))...)))).....))).))))). ( -41.60) >DroSim_CAF1 6112 116 - 1 ----AGUCGUCUCUGGGGAGAAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAAAUGCGCCGUUACAUUAACUUUUACAGGGUGUCUGCCCCAA ----.........((((((((.(((((((((.(((((((((.....)))((((((.....))))))..))))))........)))))((((...))))...)))).....))).))))). ( -41.60) >DroEre_CAF1 19730 113 - 1 -------AGUCUCUGGGGAGCAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAAAUGCGCCGUUACAUUAACUUUUACAGGGUGUCUGCCCCAA -------......((((((((((.(.(((((.(((((((((.....)))((((((.....))))))..))))))........)))))((((...)))).......).))).)).))))). ( -40.60) >DroYak_CAF1 11321 120 - 1 AAAAUACCGUCUCUGGGGAGAAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAAAUACGCCGUUACAUUAACUUUUACAGGGUGUCUGCCCCAA .............((((((((.(((((((((((((((((((.....)))((((((.....))))))..)))))).......))))))((((...))))...)))).....))).))))). ( -42.21) >consensus ____AGUCGUCUCUGGGGAGAAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAAAUGCGCCGUUACAUUAACUUUUACAGGGUGUCUGCCCCAA ..............((((..(((((((((((.(((((((((.....)))((((((.....))))))..))))))........)))))..))))))........(((.....))))))).. (-38.82 = -39.06 + 0.24)

| Location | 24,779,693 – 24,779,790 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -36.94 |
| Consensus MFE | -30.16 |
| Energy contribution | -30.56 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24779693 97 - 27905053 GUCCGUU-U-GUUGGCAG-----GCGAAAAA-------------AGU---CUCCGGGGAGAAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAA (((((((-(-(....)))-----))......-------------..(---(((....))))....))))...(((((((((.....)))((((((.....))))))..))))))...... ( -36.90) >DroSec_CAF1 10775 106 - 1 GUCCGUU-UAGCUGGCAGUCGUGGCGAAAAA-------------AGUCGUCUCUGGGGAGAAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAA .......-..(((.(((.((..(((((....-------------..)))))((....)))).))).)))...(((((((((.....)))((((((.....))))))..))))))...... ( -36.10) >DroSim_CAF1 6152 101 - 1 GUCCGUU-UUGUUGGCAG-----GCGAAAAA-------------AGUCGUCUCUGGGGAGAAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAA (((((((-((.(..(.((-----((((....-------------..)))))))..).))))...)))))...(((((((((.....)))((((((.....))))))..))))))...... ( -39.50) >DroEre_CAF1 19770 97 - 1 GUCCGUU--UGUUGGCAG-----GCGAAAAA----------------AGUCUCUGGGGAGCAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAA (((((((--(.(..(.((-----((......----------------.)))))..).))))....))))...(((((((((.....)))((((((.....))))))..))))))...... ( -36.50) >DroYak_CAF1 11361 115 - 1 GUCCGUUUUUGUUGGCAG-----GCACAAAACCAAAAAAAAAAAUACCGUCUCUGGGGAGAAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAA ((((((((((.(..(.((-----((.......................)))))..).))))))..))))...(((((((((.....)))((((((.....))))))..))))))...... ( -35.70) >consensus GUCCGUU_UUGUUGGCAG_____GCGAAAAA_____________AGUCGUCUCUGGGGAGAAUGUGGGCGUGGCAUGCGGCCGAGUGCCGGAGUGGAAUUCAUUCCGUGCGUGCAAUUAA (((((.........((.......))........................((((....))))...)))))...(((((((((.....)))((((((.....))))))..))))))...... (-30.16 = -30.56 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:31:20 2006