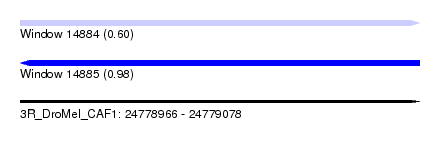
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,778,966 – 24,779,078 |
| Length | 112 |
| Max. P | 0.979713 |

| Location | 24,778,966 – 24,779,078 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.37 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -13.40 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24778966 112 + 27905053 GCGGGCCAAAAGGGGU----AAAUUGGUAGAAAGUUGGCAGACAUCCUUGAAUUUCCAUGCAAUUUUGGUAUUCCUUUUUUUCAUUUCACACCAGG----UUUUCCCUUUGGCAAAGAUU ....((((((.((((.----...(((((.(((((((....)))......((((..(((........))).))))..........))))..))))).----...))))))))))....... ( -26.90) >DroSec_CAF1 10070 101 + 1 GCGGGCCAAAAGGGGUAAGUAAAUUGGCAGAAAGUUGGCAGACUUCCUUGAAUUUGGAUGCAAUU---------------UUCACUUCACACCUAC----UUUUCCUUUUGGCUGAGAUU ...((((((((((((.(((((...(((..((((((((.((....(((........))))))))))---------------)))...)))....)))----))))))))))))))...... ( -30.50) >DroSim_CAF1 5446 101 + 1 GCGGGCCAAAAGGGGUAAGUAAAUUGGCAGAAAGUUGGCAGACUUCCUUGAAUUUGGAUGCAAUU---------------UUCACUUCACACCUAC----UUUUCCUUUUGGCUGAGUUU ...((((((((((((.(((((...(((..((((((((.((....(((........))))))))))---------------)))...)))....)))----))))))))))))))...... ( -30.50) >DroEre_CAF1 19044 95 + 1 GCGGGCCAAAAGGGGU----AAAUUUGGAGAAAGUUGGCAGACGUCCUUGAAUUUGCAUGGAA---------------------AUUCCUGCACUCCUUCUUUUCCAUUUGGCUGAGAUU ...(((((((.((((.----......((((......(((....)))........((((.((..---------------------...)))))))))).....)))).)))))))...... ( -25.90) >DroYak_CAF1 10624 107 + 1 UCGAGCCAAAAGGGUU----AAAUUGGCAGAAAGUUGGCAGAGUUCCCUGAAUUUCCAUGCAU---------UUCUUAUUUUCAUUUUAUACCUGCUUUUUUUUCCUUUUGGCUAAGAUU ...(((((((((((..----(((..(((((...(((((..((((((...))))))))).))..---------....(((.........))).)))))..))).)))))))))))...... ( -27.10) >consensus GCGGGCCAAAAGGGGU____AAAUUGGCAGAAAGUUGGCAGACUUCCUUGAAUUUGCAUGCAAUU_______________UUCACUUCACACCUAC____UUUUCCUUUUGGCUGAGAUU ...(((((((((((.........(..((.....))..)(((......))).....................................................)))))))))))...... (-13.40 = -13.80 + 0.40)


| Location | 24,778,966 – 24,779,078 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.37 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -15.52 |
| Energy contribution | -16.60 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24778966 112 - 27905053 AAUCUUUGCCAAAGGGAAAA----CCUGGUGUGAAAUGAAAAAAAGGAAUACCAAAAUUGCAUGGAAAUUCAAGGAUGUCUGCCAACUUUCUACCAAUUU----ACCCCUUUUGGCCCGC .......(((((((((....----..((((((................))))))((((((..((((((.....((....))......)))))).))))))----...))))))))).... ( -24.89) >DroSec_CAF1 10070 101 - 1 AAUCUCAGCCAAAAGGAAAA----GUAGGUGUGAAGUGAA---------------AAUUGCAUCCAAAUUCAAGGAAGUCUGCCAACUUUCUGCCAAUUUACUUACCCCUUUUGGCCCGC .......(((((((((..((----(((((((..(......---------------..)..))))........(((((((......))))))).......)))))...))))))))).... ( -27.30) >DroSim_CAF1 5446 101 - 1 AAACUCAGCCAAAAGGAAAA----GUAGGUGUGAAGUGAA---------------AAUUGCAUCCAAAUUCAAGGAAGUCUGCCAACUUUCUGCCAAUUUACUUACCCCUUUUGGCCCGC .......(((((((((..((----(((((((..(......---------------..)..))))........(((((((......))))))).......)))))...))))))))).... ( -27.30) >DroEre_CAF1 19044 95 - 1 AAUCUCAGCCAAAUGGAAAAGAAGGAGUGCAGGAAU---------------------UUCCAUGCAAAUUCAAGGACGUCUGCCAACUUUCUCCAAAUUU----ACCCCUUUUGGCCCGC .......((((((.((.(((...((((.(((((...---------------------.(((.((......)).)))..))))).......))))...)))----.))...)))))).... ( -22.10) >DroYak_CAF1 10624 107 - 1 AAUCUUAGCCAAAAGGAAAAAAAAGCAGGUAUAAAAUGAAAAUAAGAA---------AUGCAUGGAAAUUCAGGGAACUCUGCCAACUUUCUGCCAAUUU----AACCCUUUUGGCUCGA ......((((((((((........((((((((...((....)).....---------)))).(((.....((((....))))))).....))))......----...))))))))))... ( -22.83) >consensus AAUCUCAGCCAAAAGGAAAA____GCAGGUGUGAAAUGAA_______________AAUUGCAUGCAAAUUCAAGGAAGUCUGCCAACUUUCUGCCAAUUU____ACCCCUUUUGGCCCGC .......(((((((((...........(((((((.......................))))))).(((((..(((((((......)))))))...))))).......))))))))).... (-15.52 = -16.60 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:31:17 2006