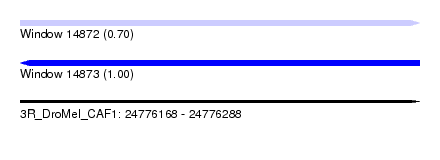
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,776,168 – 24,776,288 |
| Length | 120 |
| Max. P | 0.995809 |

| Location | 24,776,168 – 24,776,288 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -43.12 |
| Consensus MFE | -37.60 |
| Energy contribution | -37.92 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24776168 120 + 27905053 ACAAAUUCGUAAGACCGCUGGUGCCGUAUGACCUUUGACCCAGCUGCUACCGCGUUGAGCAGAAACAGCGACAGUAGCCAAAUCGGCAGCUGCUGCUUGGCGGUCAAAGUUGCGGAUGAG ....((((((((.((.((....)).)).....((((((((.....((....))((..(((((.........((((.(((.....))).)))))))))..))))))))))))))))))... ( -42.00) >DroSec_CAF1 7338 120 + 1 ACAAAUUCGCAAGACCGCUGGUGCCGUAUGACCUUUGACCCAGCUGCUACCGCGUUGAGCAGAAACAGCGACAGUAGCCAAAUCGGCAGCUGCUGCUUGGCGGUCAAAGUUGCGGAUGAG ....((((((((.((.((....)).)).....((((((((.....((....))((..(((((.........((((.(((.....))).)))))))))..))))))))))))))))))... ( -44.00) >DroSim_CAF1 2736 120 + 1 ACAAAUUCGUAUGAACGCUGGUGCCGUAUGACCUUUGACCCAGCUGCUACCGCGUUGAGCAGAAACAGCGACAGUAGCCAAAUCGGCAGCUGCUGCUUGGCGGUCAAAGUUGCGGAUGAG .................((.((.(((((....((((((((.....((....))((..(((((.........((((.(((.....))).)))))))))..)))))))))).))))))).)) ( -40.40) >DroEre_CAF1 16251 120 + 1 ACAUAUUCGUAAGACCGCUGGUGCCGUAUGACCUUUGACCCAGCUGCUACCGCGCUGACCAGAAACAGCGACAGUAGCCAAAUCGGCAGCUGCUGCUUGGCGGUCAAAGUUGCGGAUGAG ...(((((((((.((.((....)).)).....((((((((((((.((....)))))).(((.....((((.((((.(((.....))).)))).))))))).))))))))))))))))).. ( -43.80) >DroYak_CAF1 7825 120 + 1 ACAUAUGCGUAAGUUGGCCAGUGCCGUGUGACCUUUGACCCAGCUGCUACCGCGUUGGCCAGAAACAGCGACAGUAGCCAAAUCGGCAGCUGCUGCUUGGCGGUCAAAGUUGCGGAUGAG ...(((.(((((...(((....))).......((((((((((((.((....))))))((((.....((((.((((.(((.....))).)))).))))))))))))))))))))).))).. ( -45.40) >consensus ACAAAUUCGUAAGACCGCUGGUGCCGUAUGACCUUUGACCCAGCUGCUACCGCGUUGAGCAGAAACAGCGACAGUAGCCAAAUCGGCAGCUGCUGCUUGGCGGUCAAAGUUGCGGAUGAG ....((((((((....((....))........((((((((.....((....))((..(((((.........((((.(((.....))).)))))))))..))))))))))))))))))... (-37.60 = -37.92 + 0.32)

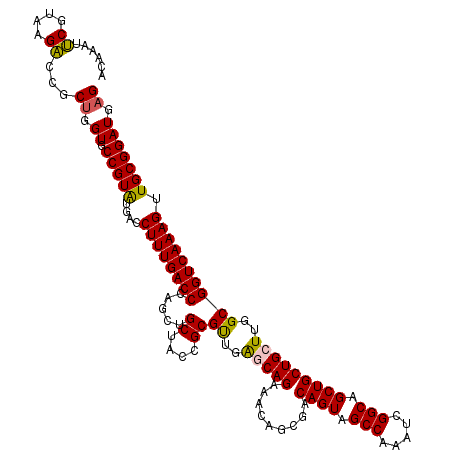

| Location | 24,776,168 – 24,776,288 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -44.54 |
| Consensus MFE | -39.76 |
| Energy contribution | -40.76 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24776168 120 - 27905053 CUCAUCCGCAACUUUGACCGCCAAGCAGCAGCUGCCGAUUUGGCUACUGUCGCUGUUUCUGCUCAACGCGGUAGCAGCUGGGUCAAAGGUCAUACGGCACCAGCGGUCUUACGAAUUUGU .....((((..(((((((((((((((.((....)).).)))))).......((((((.((((.....)))).))))))..))))))))(((....)))....)))).............. ( -45.00) >DroSec_CAF1 7338 120 - 1 CUCAUCCGCAACUUUGACCGCCAAGCAGCAGCUGCCGAUUUGGCUACUGUCGCUGUUUCUGCUCAACGCGGUAGCAGCUGGGUCAAAGGUCAUACGGCACCAGCGGUCUUGCGAAUUUGU (.((.((((..(((((((((((((((.((....)).).)))))).......((((((.((((.....)))).))))))..))))))))(((....)))....))))...)).)....... ( -45.50) >DroSim_CAF1 2736 120 - 1 CUCAUCCGCAACUUUGACCGCCAAGCAGCAGCUGCCGAUUUGGCUACUGUCGCUGUUUCUGCUCAACGCGGUAGCAGCUGGGUCAAAGGUCAUACGGCACCAGCGUUCAUACGAAUUUGU ......(((..(((((((((((((((.((....)).).)))))).......((((((.((((.....)))).))))))..))))))))(((....)))....)))............... ( -41.90) >DroEre_CAF1 16251 120 - 1 CUCAUCCGCAACUUUGACCGCCAAGCAGCAGCUGCCGAUUUGGCUACUGUCGCUGUUUCUGGUCAGCGCGGUAGCAGCUGGGUCAAAGGUCAUACGGCACCAGCGGUCUUACGAAUAUGU .....((((..((((((((......((((.(((((((....(((....)))((((........)))).))))))).))))))))))))(((....)))....)))).............. ( -45.50) >DroYak_CAF1 7825 120 - 1 CUCAUCCGCAACUUUGACCGCCAAGCAGCAGCUGCCGAUUUGGCUACUGUCGCUGUUUCUGGCCAACGCGGUAGCAGCUGGGUCAAAGGUCACACGGCACUGGCCAACUUACGCAUAUGU .......((..((((((((......((((.(((((((..(((((((..(.......)..)))))))..))))))).)))))))))))).......(((....))).......))...... ( -44.80) >consensus CUCAUCCGCAACUUUGACCGCCAAGCAGCAGCUGCCGAUUUGGCUACUGUCGCUGUUUCUGCUCAACGCGGUAGCAGCUGGGUCAAAGGUCAUACGGCACCAGCGGUCUUACGAAUUUGU .....((((..(((((((((((((((.((....)).).)))))).......((((((.((((.....)))).))))))..))))))))(((....)))....)))).............. (-39.76 = -40.76 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:31:07 2006