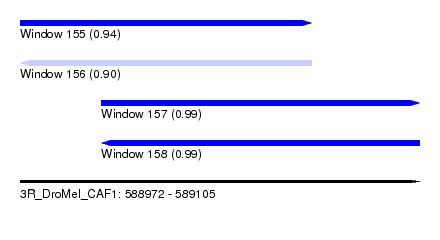
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 588,972 – 589,105 |
| Length | 133 |
| Max. P | 0.991435 |

| Location | 588,972 – 589,069 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.47 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -17.52 |
| Energy contribution | -18.42 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 588972 97 + 27905053 AUCGGGGCCAUUAAAGC------CUAUA-------UUCGAGUUAGCGU-CAUCACGAUAUCGUGACCAGACGUUGAGUCAGACGAAAGCUAGCAUAAAUAUUUGAUUAGUU .(((((((.......))------))...-------...((.(((((((-(.(((((....)))))...)))))))).))...))).((((((((........)).)))))) ( -29.70) >DroSec_CAF1 5455 104 + 1 AUCGUGGUCAUUAAAGU------CAAUAUGAGCUGUUCGAGUCAGCGU-CGUCACGAUAUCGUGACCAGACGCUCAGCCAGACGAAAGCUAGCAUAAAUAUUUGAUUAGUU .(((((..(......).------...)))))((((.((((((.(((((-(((((((....))))))..))))))..((.((.(....))).))......)))))).)))). ( -30.00) >DroSim_CAF1 4218 104 + 1 AUCGUGGUCAUUAAAGU------CAAUAUGAGCUGUUCGAGUUAGCGU-CGUCACGAUAUCGUGACCAGACGCUCAGCCAGACGAAAGCUAGCAUAAAUAUUUGAUUAGUU .(((((..(......).------...)))))((((.((((((.(((((-(((((((....))))))..))))))..((.((.(....))).))......)))))).)))). ( -29.90) >DroEre_CAF1 6469 88 + 1 ----------------C------CAAUAUGAGCUAUUUGAAUUAGCGU-AGUCACGACAUCGUGACCAGACGCAGAGCCAGACUUGUACUAGCAUAAAUAUUUGAUGAGUU ----------------.------........((((((((..((.((((-.((((((....))))))...)))).))..)))).......)))).................. ( -20.21) >DroYak_CAF1 6920 104 + 1 AUUGGGGCUAUUGAAGC------CUAUAUGACCUUUUCGAGUUAGCGG-UGUCACAACAUUGUGACCAGACGCAGACCCAGACAAAAACUGGAAUAAAUAUUUGACGAGUU ....(((((.....)))------)).......(((.(((((((.(((.-.((((((....))))))....))).)))((((.......)))).........)))).))).. ( -27.10) >DroAna_CAF1 6527 110 + 1 UUUGGCACUACUA-UGCGUACUAUAAUAUGAGCUAUUCGAGUUAACGUUUGUCACGAUAUCGUGACCAGACGUCCAGCCAGACGAAAGCGAGCAUAAAUAUUUGAUUAGUU ....(((......-))).............(((((.((((((..((((((((((((....))))).)))))))...((..(.(....))..))......)))))).))))) ( -28.40) >consensus AUCGGGGCCAUUAAAGC______CAAUAUGAGCUAUUCGAGUUAGCGU_CGUCACGAUAUCGUGACCAGACGCUCAGCCAGACGAAAGCUAGCAUAAAUAUUUGAUUAGUU ..............................(((((.((((((..((((..((((((....))))))...))))...((.((.(....))).))......)))))).))))) (-17.52 = -18.42 + 0.89)

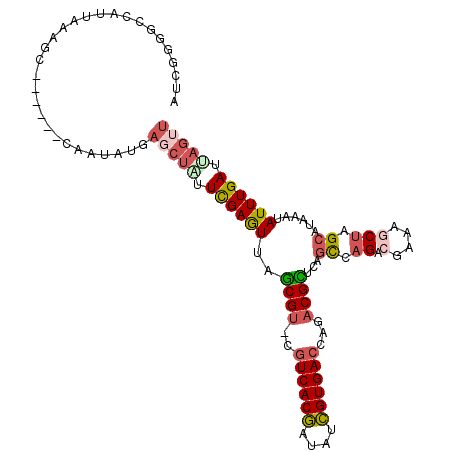
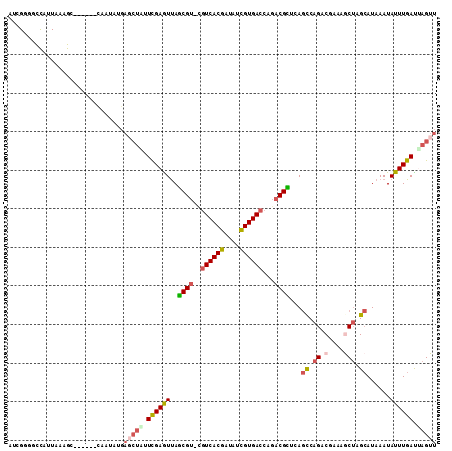
| Location | 588,972 – 589,069 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.47 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.13 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 588972 97 - 27905053 AACUAAUCAAAUAUUUAUGCUAGCUUUCGUCUGACUCAACGUCUGGUCACGAUAUCGUGAUG-ACGCUAACUCGAA-------UAUAG------GCUUUAAUGGCCCCGAU .....(((..((((((..(.((((....(((.(((.....)))..((((((....)))))))-)))))).)..)))-------))).(------(((.....))))..))) ( -23.20) >DroSec_CAF1 5455 104 - 1 AACUAAUCAAAUAUUUAUGCUAGCUUUCGUCUGGCUGAGCGUCUGGUCACGAUAUCGUGACG-ACGCUGACUCGAACAGCUCAUAUUG------ACUUUAAUGACCACGAU ......((((.....((((..((((((((...((.(.((((((..((((((....)))))))-))))).))))))).)))))))))))------)................ ( -27.00) >DroSim_CAF1 4218 104 - 1 AACUAAUCAAAUAUUUAUGCUAGCUUUCGUCUGGCUGAGCGUCUGGUCACGAUAUCGUGACG-ACGCUAACUCGAACAGCUCAUAUUG------ACUUUAAUGACCACGAU ......((((.....((((..((((((((...(....((((((..((((((....)))))))-)))))..).)))).)))))))))))------)................ ( -27.00) >DroEre_CAF1 6469 88 - 1 AACUCAUCAAAUAUUUAUGCUAGUACAAGUCUGGCUCUGCGUCUGGUCACGAUGUCGUGACU-ACGCUAAUUCAAAUAGCUCAUAUUG------G---------------- ...........(((((..(((((.......)))))...((((..(((((((....)))))))-))))......)))))..........------.---------------- ( -21.40) >DroYak_CAF1 6920 104 - 1 AACUCGUCAAAUAUUUAUUCCAGUUUUUGUCUGGGUCUGCGUCUGGUCACAAUGUUGUGACA-CCGCUAACUCGAAAAGGUCAUAUAG------GCUUCAAUAGCCCCAAU ..((..((...........((((.......))))((..(((....((((((....)))))).-.)))..))..))..))........(------(((.....))))..... ( -24.40) >DroAna_CAF1 6527 110 - 1 AACUAAUCAAAUAUUUAUGCUCGCUUUCGUCUGGCUGGACGUCUGGUCACGAUAUCGUGACAAACGUUAACUCGAAUAGCUCAUAUUAUAGUACGCA-UAGUAGUGCCAAA ...............((((...(((((((...(....(((((...((((((....))))))..)))))..).)))).))).)))).....((((...-.....)))).... ( -24.90) >consensus AACUAAUCAAAUAUUUAUGCUAGCUUUCGUCUGGCUCAGCGUCUGGUCACGAUAUCGUGACG_ACGCUAACUCGAACAGCUCAUAUUG______GCUUUAAUGGCCCCGAU ..................((((((....).)))))..(((((...((((((....))))))..)))))........................................... (-16.49 = -16.13 + -0.36)


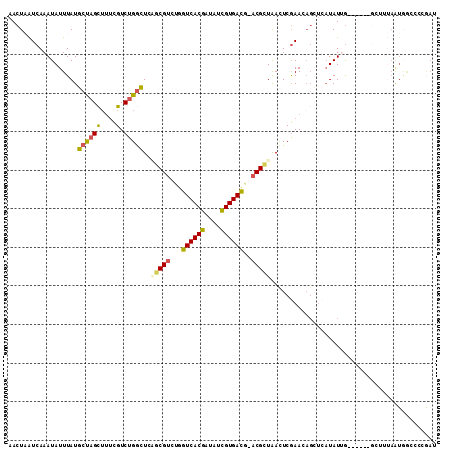
| Location | 588,999 – 589,105 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.23 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -18.02 |
| Energy contribution | -19.25 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 588999 106 + 27905053 GUUAGCGU-CAUCACGAUAUCGUGACCAGACGUUGAGUCAGACGAAAGCUAGCAUAAAUAUUUGAUUAGUUUCGCUGGCCCCGCAGUUUUCGCCCCCGCCUGCC--UUU-- .(((((((-(.(((((....)))))...))))))))(((((.(((((.((((((........)).))))))))))))))...((((.....((....)))))).--...-- ( -32.60) >DroSec_CAF1 5489 82 + 1 GUCAGCGU-CGUCACGAUAUCGUGACCAGACGCUCAGCCAGACGAAAGCUAGCAUAAAUAUUUGAUUAGUUUCACUGGCCUCG---------------------------- ...(((((-(((((((....))))))..))))))..(((((..((((.((((((........)).)))))))).)))))....---------------------------- ( -30.00) >DroSim_CAF1 4252 82 + 1 GUUAGCGU-CGUCACGAUAUCGUGACCAGACGCUCAGCCAGACGAAAGCUAGCAUAAAUAUUUGAUUAGUUUCACUGGCCUCG---------------------------- ...(((((-(((((((....))))))..))))))..(((((..((((.((((((........)).)))))))).)))))....---------------------------- ( -29.90) >DroEre_CAF1 6487 81 + 1 AUUAGCGU-AGUCACGACAUCGUGACCAGACGCAGAGCCAGACUUGUACUAGCAUAAAUAUUUGAUGAGUUGCGCCUGCCUC----------------------------- ....((((-.((((((....))))))...)))).(((.(((...((((((..((........))...)).)))).))).)))----------------------------- ( -22.70) >DroYak_CAF1 6954 82 + 1 GUUAGCGG-UGUCACAACAUUGUGACCAGACGCAGACCCAGACAAAAACUGGAAUAAAUAUUUGACGAGUUUUGCUGGCCCCG---------------------------- (((.(((.-.((((((....))))))....))).)))((((...((((((.(.............).)))))).)))).....---------------------------- ( -22.12) >DroAna_CAF1 6566 103 + 1 GUUAACGUUUGUCACGAUAUCGUGACCAGACGUCCAGCCAGACGAAAGCGAGCAUAAAUAUUUGAUUAGUUUGGCCGAACCGGGA--------CUCUGACUGGCACUUUGC ((((......((((((....))))))((((.((((.(((((((.....((((........))))....)))))))((...)))))--------)))))..))))....... ( -30.70) >consensus GUUAGCGU_CGUCACGAUAUCGUGACCAGACGCUCAGCCAGACGAAAGCUAGCAUAAAUAUUUGAUUAGUUUCGCUGGCCCCG____________________________ ....((((..((((((....))))))...))))...(((((....(((((((((........)).)))))))..)))))................................ (-18.02 = -19.25 + 1.23)

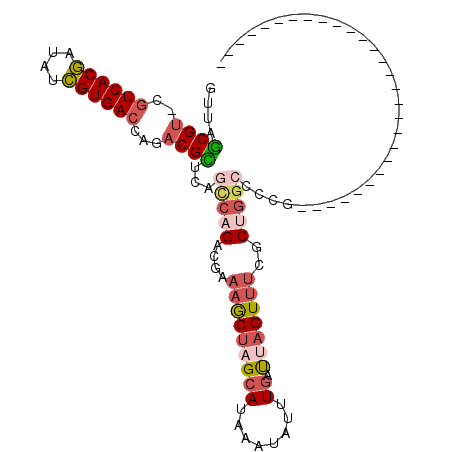
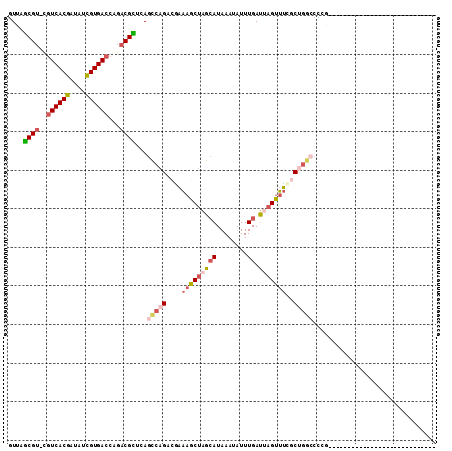
| Location | 588,999 – 589,105 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.23 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -17.95 |
| Energy contribution | -19.18 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 588999 106 - 27905053 --AAA--GGCAGGCGGGGGCGAAAACUGCGGGGCCAGCGAAACUAAUCAAAUAUUUAUGCUAGCUUUCGUCUGACUCAACGUCUGGUCACGAUAUCGUGAUG-ACGCUAAC --...--((((((((..(((.......((.......))((......))..............)))..)))))).))...((((..((((((....)))))))-)))..... ( -30.80) >DroSec_CAF1 5489 82 - 1 ----------------------------CGAGGCCAGUGAAACUAAUCAAAUAUUUAUGCUAGCUUUCGUCUGGCUGAGCGUCUGGUCACGAUAUCGUGACG-ACGCUGAC ----------------------------...((((((((((((((..((........)).))).))))).)))))).((((((..((((((....)))))))-)))))... ( -31.50) >DroSim_CAF1 4252 82 - 1 ----------------------------CGAGGCCAGUGAAACUAAUCAAAUAUUUAUGCUAGCUUUCGUCUGGCUGAGCGUCUGGUCACGAUAUCGUGACG-ACGCUAAC ----------------------------...((((((((((((((..((........)).))).))))).)))))).((((((..((((((....)))))))-)))))... ( -31.50) >DroEre_CAF1 6487 81 - 1 -----------------------------GAGGCAGGCGCAACUCAUCAAAUAUUUAUGCUAGUACAAGUCUGGCUCUGCGUCUGGUCACGAUGUCGUGACU-ACGCUAAU -----------------------------(((.(((((...(((...((........))..)))....))))).))).((((..(((((((....)))))))-)))).... ( -27.10) >DroYak_CAF1 6954 82 - 1 ----------------------------CGGGGCCAGCAAAACUCGUCAAAUAUUUAUUCCAGUUUUUGUCUGGGUCUGCGUCUGGUCACAAUGUUGUGACA-CCGCUAAC ----------------------------.(((.(((((((((((.(.............).)).))))).)))).)))(((....((((((....)))))).-.))).... ( -21.92) >DroAna_CAF1 6566 103 - 1 GCAAAGUGCCAGUCAGAG--------UCCCGGUUCGGCCAAACUAAUCAAAUAUUUAUGCUCGCUUUCGUCUGGCUGGACGUCUGGUCACGAUAUCGUGACAAACGUUAAC ........(((((((((.--------...(((...(((.....(((........))).....))).))))))))))))((((...((((((....))))))..)))).... ( -28.50) >consensus ____________________________CGAGGCCAGCGAAACUAAUCAAAUAUUUAUGCUAGCUUUCGUCUGGCUCAGCGUCUGGUCACGAUAUCGUGACG_ACGCUAAC ...............................((((((((((((((...............))).))))).)))))).(((((...((((((....))))))..)))))... (-17.95 = -19.18 + 1.23)

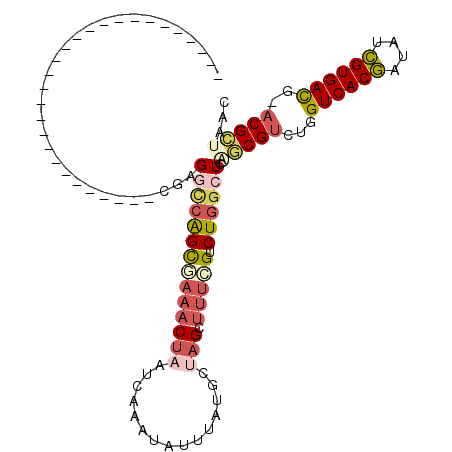
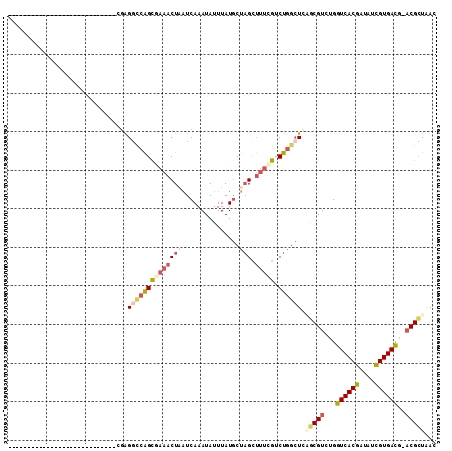
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:54 2006