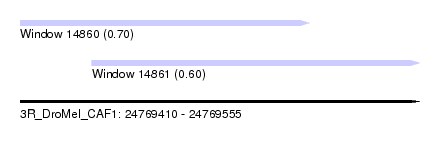
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,769,410 – 24,769,555 |
| Length | 145 |
| Max. P | 0.700644 |

| Location | 24,769,410 – 24,769,515 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.56 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
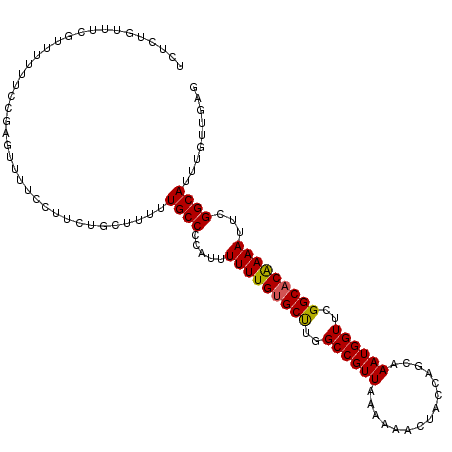
>3R_DroMel_CAF1 24769410 105 + 27905053 --UCUGUAUGUUUUUUU-----UUUUUUAGCCCAUUUUUGCCCCAUUUUUUGCGCCUGGCCGUUAAAAAACUACAAGCAAAUGGUUCGGCACAAAAUUCGGCAUUUGUUCUG --...............-----.....(((..((....((((.....(((((.(((..((((((...............))))))..))).)))))...))))..))..))) ( -22.06) >DroPse_CAF1 21418 112 + 1 UCUCUGUUUCGUUUUUUCCGAGUUUUCCUUCUGCUUUUUGCCCCAUUUUUUGUGCUUGGCCGUUAAAAAACUACCAGCAAAUGGUUCGGCACAAAAUUCGGCAUUUGUUGAG .......((((.......)))).......((.((....((((.....(((((((((..((((((...............))))))..)))))))))...))))...)).)). ( -25.26) >DroPer_CAF1 21712 112 + 1 UCUCUGUUUCGUUUUUUCCGAGUUUUCCUUCUGCUUUUUGCCCCAUUUUUUGUGCUUGGCCGUUAAAAAACUACCAGCAAAUGGUUCGGCACGAAAUUCGGCAUUUGUUGAG .......((((.......)))).......((.((....((((.....(((((((((..((((((...............))))))..)))))))))...))))...)).)). ( -24.96) >consensus UCUCUGUUUCGUUUUUUCCGAGUUUUCCUUCUGCUUUUUGCCCCAUUUUUUGUGCUUGGCCGUUAAAAAACUACCAGCAAAUGGUUCGGCACAAAAUUCGGCAUUUGUUGAG ......................................((((.....(((((((((..((((((...............))))))..)))))))))...))))......... (-21.67 = -21.56 + -0.11)


| Location | 24,769,436 – 24,769,555 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.56 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
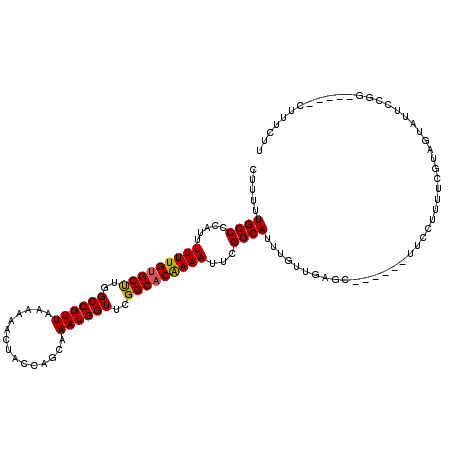
>3R_DroMel_CAF1 24769436 119 + 27905053 AUUUUUGCCCCAUUUUUUGCGCCUGGCCGUUAAAAAACUACAAGCAAAUGGUUCGGCACAAAAUUCGGCAUUUGUUCUGCCUCUAUUUCCUUGUCUUUGUUUUCCGGGCCCUCGUUUUU ......((((....(((((.(((..((((((...............))))))..))).)))))...((((.......))))........................)))).......... ( -26.96) >DroPse_CAF1 21451 108 + 1 CUUUUUGCCCCAUUUUUUGUGCUUGGCCGUUAAAAAACUACCAGCAAAUGGUUCGGCACAAAAUUCGGCAUUUGUUGAGC------UUCCUUUUCGUAGUAUUCCGG-----CUUUCUU ......(((.....(((((((((..((((((...............))))))..)))))))))((((((....)))))).------...................))-----)...... ( -25.26) >DroPer_CAF1 21745 108 + 1 CUUUUUGCCCCAUUUUUUGUGCUUGGCCGUUAAAAAACUACCAGCAAAUGGUUCGGCACGAAAUUCGGCAUUUGUUGAGC------UUCCUUUUCGUAGUAUUCCGG-----CUUUCUU ......(((.....(((((((((..((((((...............))))))..)))))))))((((((....)))))).------...................))-----)...... ( -24.96) >consensus CUUUUUGCCCCAUUUUUUGUGCUUGGCCGUUAAAAAACUACCAGCAAAUGGUUCGGCACAAAAUUCGGCAUUUGUUGAGC______UUCCUUUUCGUAGUAUUCCGG_____CUUUCUU .....((((.....(((((((((..((((((...............))))))..)))))))))...))))................................................. (-21.67 = -21.56 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:30:56 2006