
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,748,927 – 24,749,087 |
| Length | 160 |
| Max. P | 0.991416 |

| Location | 24,748,927 – 24,749,047 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.97 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24748927 120 - 27905053 ACAUAGGACGUAGGAUCAGCGGGUCCCAGCGCCCUCCUGGUCAUCCACUCCUCCUGGAAACUAUUAUAUAUCGAUGCUUACAAUUGUUGCUAAUAAGUUAAACAAAUCACCUACGUCCGC .....(((((((((...(((((((.((((.......))))..))))..(((....))).................))).....((((((((....)))..)))))....))))))))).. ( -33.30) >DroSec_CAF1 2973 118 - 1 ACAUAGGGCGUAGGAUCAGCGGGUCCUAGCGCCCUCCUGGUCAUCCACUCCUUCUGGAAACUAUUA--UAUCGAUGCUUACAAUGGUAGCUAAUAAGUUAAACAAAUCACCCACGUUCGC ....(((((((((((((....)))))).)))))))..(((...((((.......))))((((((((--(((((.((....)).)))))..)))).))))...........)))....... ( -29.20) >DroSim_CAF1 2955 118 - 1 ACAUAGGGCGCAGGAUCAGCGGGUCCUAGCGCCCUCCUGGUCAUCCACUCCUUCUGGAAACUAUUA--UAUCGAUGCUUACAAUGGUUGCUAAUGUGUUAAGCAAAUCACCUACGUCCGC ....(((((((((((((....)))))).)))))))...((.(.((((.......))))........--......((((((((.(((...)))...)).))))))..........).)).. ( -33.60) >DroEre_CAF1 2992 118 - 1 ACAUAGGGCGUAGGAUCAGCGGGCUCUAGCUCCCUCCUGGUCAUCCACUCCUGCUGGAAACUAUUA--CAUCGAUAAUUACAAUGGUUGCUAAUAAGGCAAACAAAUCACCUACGUCUUC ....((((((((((.((((((((.........((....)).........)))))))).........--...............((.(((((.....))))).)).....)))))))))). ( -32.87) >DroYak_CAF1 2980 118 - 1 ACAUAGGCCGUAGGAUCAGCGGGCCCCAGUGCCCUCCUGGUCAUCCACUCCUCCUGGAAACUAUGG--CAUCGAUGCUUACAAUGGCUGCUAAUAAGGCACACAAAUCACCUACGUCCUC ((.((((((((((((((((.((((......))))..)))))).((((.......))))..))))))--)...((((((......)))((((.....)))).....)))..))).)).... ( -36.40) >consensus ACAUAGGGCGUAGGAUCAGCGGGUCCUAGCGCCCUCCUGGUCAUCCACUCCUCCUGGAAACUAUUA__UAUCGAUGCUUACAAUGGUUGCUAAUAAGUUAAACAAAUCACCUACGUCCGC .....((((((((((((((.((((......))))..)))))).((((.......))))....................................................)))))))).. (-26.20 = -26.44 + 0.24)

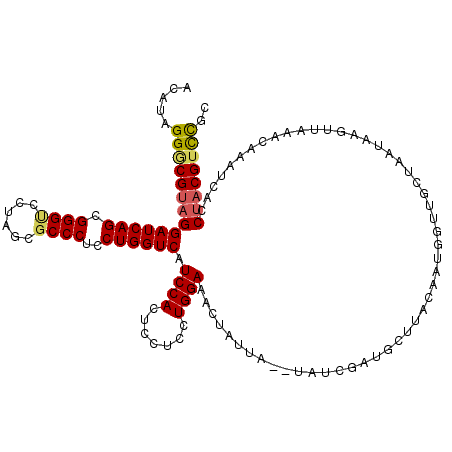

| Location | 24,748,967 – 24,749,087 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -41.16 |
| Consensus MFE | -34.58 |
| Energy contribution | -35.86 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24748967 120 - 27905053 CGUGCGUCGCAUGAGCGGAAAACUUAUCAACGGAUUAGCCACAUAGGACGUAGGAUCAGCGGGUCCCAGCGCCCUCCUGGUCAUCCACUCCUCCUGGAAACUAUUAUAUAUCGAUGCUUA ...((((((...(((.(((..........(((..(((......)))..)))..((((((.((((......))))..)))))).))).))).....(....)..........))))))... ( -33.40) >DroSec_CAF1 3013 118 - 1 CAUGCGUCGCAUGAGCGGAAAGCUUAUCAACGGAUUGGCCACAUAGGGCGUAGGAUCAGCGGGUCCUAGCGCCCUCCUGGUCAUCCACUCCUUCUGGAAACUAUUA--UAUCGAUGCUUA ...((((((.((((((.....))))))....((((.(((((...(((((((((((((....)))))).)))))))..)))))))))..(((....)))........--...))))))... ( -49.10) >DroSim_CAF1 2995 118 - 1 CAUGCGUCGCAUGAGCGGAAAGCUUAUCAACGGAUUGGCCACAUAGGGCGCAGGAUCAGCGGGUCCUAGCGCCCUCCUGGUCAUCCACUCCUUCUGGAAACUAUUA--UAUCGAUGCUUA ...((((((.((((((.....))))))....((((.(((((...(((((((((((((....)))))).)))))))..)))))))))..(((....)))........--...))))))... ( -51.10) >DroEre_CAF1 3032 118 - 1 CAUACGUCGCAUGAGCGGAAAACUUAUCAACGGAUUGGCCACAUAGGGCGUAGGAUCAGCGGGCUCUAGCUCCCUCCUGGUCAUCCACUCCUGCUGGAAACUAUUA--CAUCGAUAAUUA .....((((((.(((.(((......(((....)))((((((...((((.((.(((((....)).))).)).))))..))))))))).))).))).(....).....--....)))..... ( -31.30) >DroYak_CAF1 3020 118 - 1 CAUGCGUCGCAUGAGCGGAAAACUUAUCAAAGGAUUGGCCACAUAGGCCGUAGGAUCAGCGGGCCCCAGUGCCCUCCUGGUCAUCCACUCCUCCUGGAAACUAUGG--CAUCGAUGCUUA ...((((((..((.((.((...(((....)))..)).)))).....(((((((((((((.((((......))))..)))))).((((.......))))..))))))--)..))))))... ( -40.90) >consensus CAUGCGUCGCAUGAGCGGAAAACUUAUCAACGGAUUGGCCACAUAGGGCGUAGGAUCAGCGGGUCCUAGCGCCCUCCUGGUCAUCCACUCCUCCUGGAAACUAUUA__UAUCGAUGCUUA ...((((((.(((..................((((.(((((...(((((((((((((....)))))).)))))))..)))))))))..(((....)))..........)))))))))... (-34.58 = -35.86 + 1.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:30:44 2006