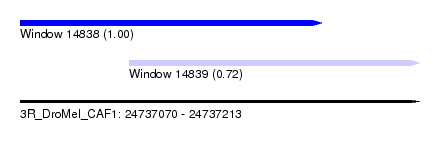
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,737,070 – 24,737,213 |
| Length | 143 |
| Max. P | 0.997304 |

| Location | 24,737,070 – 24,737,178 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 96.03 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -29.58 |
| Energy contribution | -29.98 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24737070 108 + 27905053 CGGUUUCUGUACUUUCCCUCGCUUUGCUAAUUUCUGUUG-GUUGGCCCCCAAGUUUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAGAGCAG .((.....((((........(((..((((((....))))-)).)))..........))))..))....(((((((..((((((((....))))))))..)))))))... ( -33.47) >DroSec_CAF1 11626 108 + 1 CGUUUUCUGUACUUUCCCUCGCUUUGCUAAUUUCUGUUG-GUUGGCCCCCAAGUUUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAUAGCAG .................................((((((-(...(((.(((((...((((.....)))).))))).)))((((((....))))))....)))))))... ( -29.90) >DroSim_CAF1 11984 108 + 1 CGUUUUCUGUACUUUCCCUCGCUUUGCUAAUUUCUGUUG-GUUGGCCCCCAAGUCUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAGAGCAG ........((((........(((..((((((....))))-)).)))..........))))...(((..(((((((..((((((((....))))))))..)))))))))) ( -32.97) >DroEre_CAF1 21308 104 + 1 CGGUUUCUGUACUUUCCCUCGCUUUGCUAAUUUCAGUU-----GGCCCCCAAGUUUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAGAGCAG ......((((.(((.....(((((..(((((....)))-----)(((.(((((...((((.....)))).))))).))).........)..))))).....))).)))) ( -31.30) >DroYak_CAF1 10646 109 + 1 CGGUUUCUGUACUUUCCCUCGCUUUGCUAAUUUCAGUUGUGAUGGCCCCCAAGUUUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAGAGCAG ......((((.....((.((((...(((......))).)))).))...........((((.....))))((((((..((((((((....))))))))..)))))))))) ( -32.30) >consensus CGGUUUCUGUACUUUCCCUCGCUUUGCUAAUUUCUGUUG_GUUGGCCCCCAAGUUUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAGAGCAG ......((((.(((.....(((((..(((((....)))).....(((.(((((...((((.....)))).))))).))).........)..))))).....))).)))) (-29.58 = -29.98 + 0.40)


| Location | 24,737,109 – 24,737,213 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -28.99 |
| Consensus MFE | -20.97 |
| Energy contribution | -22.55 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24737109 104 + 27905053 -GUUGGCCCCCAAGUUUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAGAGCAGAGGCACUUAUUGCUUACCUCCAACCUUAGAAUAUC -(((((.....((((........(((.(.(((((((..((((((((....))))))))..))))))).).)))........))))....)))))........... ( -34.79) >DroSec_CAF1 11665 104 + 1 -GUUGGCCCCCAAGUUUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAUAGCAGAGGCACUUAUUGCUUACCUCCAACCUCAGAAUCUC -(((((.....((((........(((.(.((.((((..((((((((....))))))))..)))).)).).)))........))))....)))))........... ( -28.59) >DroSim_CAF1 12023 104 + 1 -GUUGGCCCCCAAGUCUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAGAGCAGAGGCACUUAUUGCUUACCUCCAACCUUAGAAUCUC -(((((.....((((..(((...(((.(.(((((((..((((((((....))))))))..))))))).).)))....))).))))....)))))........... ( -34.30) >DroEre_CAF1 21346 101 + 1 ----GGCCCCCAAGUUUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAGAGCAGAGGCACUUAUUGCUUACCUCCAACAUUAGAAUAUC ----((.....((((........(((.(.(((((((..((((((((....))))))))..))))))).).)))........))))....)).............. ( -30.49) >DroYak_CAF1 10685 105 + 1 UGAUGGCCCCCAAGUUUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAGAGCAGAGGCACUUAAUGCUUAGCUCCAACCUUAGAAUCUC .(((.(((.(((((...((((.....)))).))))).)))((((((....))))))........((((..(((((.....))))).))))..........))).. ( -31.70) >DroPer_CAF1 12670 82 + 1 ----------CGAGUUUGUACUUCCUGUACUUUU-GUGGCCUUUCAAGUGUGAAACAUCCACAGCCCCAAAAACACCUCUUUCUGAU------------GAUUGG ----------.(((...((((.....))))..((-((((..(((((....)))))...))))))............)))........------------...... ( -14.10) >consensus _GUUGGCCCCCAAGUUUGUACUUCCUGUACUCUUGGUGGCGUUUCAAGUGUGAAGCGUUCCCAAGAGCAGAGGCACUUAUUGCUUACCUCCAACCUUAGAAUCUC .....(((...((((....)))).(((..(((((((..((((((((....))))))))..)))))))))).)))............................... (-20.97 = -22.55 + 1.58)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:30:37 2006