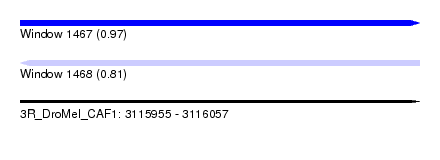
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,115,955 – 3,116,057 |
| Length | 102 |
| Max. P | 0.968280 |

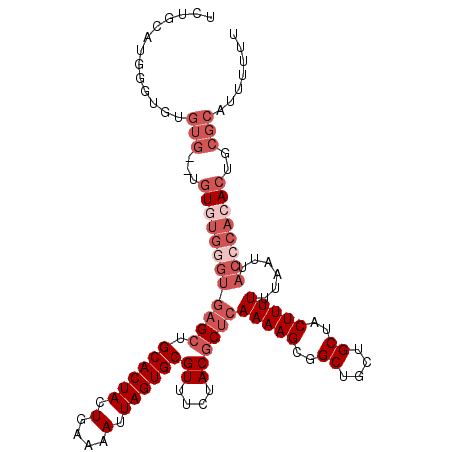
| Location | 3,115,955 – 3,116,057 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.09 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -22.16 |
| Energy contribution | -24.23 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3115955 102 + 27905053 AAAAAAUGCGCAGUGUGGGUAAUUAAAAAAGUAGCAGCAGCCGCUUUUGAGCGUAGAAACGCACUAAUUUUCAGUAGUGCAUC--------ACA--CACCCACCCAUGCAGA .........(((..((((((......(((((..((....))..)))))............((((((........))))))...--------...--.))))))...)))... ( -28.90) >DroSec_CAF1 3660 110 + 1 AAAAAAUGCGCAGUGUGGGUAAUUAAAAAAGUAGCAGCAGCCGCUUUUGAGCGUAGAAACGCACUAAUUUUCAGUAGUGCAGCUCACCCAUACA--CACACACACAUGCAGA .........(((((((((((......(((((..((....))..)))))((((((....))((((((........)))))).)))))))))))).--..........)))... ( -33.70) >DroSim_CAF1 8982 112 + 1 AAAAAAUGCGCAGUGUGGGUAAUUAAAAAAGUAGCAGCAGCCGCUUUUGAGCGUAGAAACGCACUAAUUUUCAGUAGUGCAGCUCACCCACAAAUACACACACCCCUACAGA .............(((((((......(((((..((....))..)))))((((((....))((((((........)))))).))))))))))).................... ( -31.90) >DroYak_CAF1 7181 100 + 1 AAAAAAUGCGAAGUGUGGGUAAUUAAAAAAGUAGCAGCAGCCGCUUUUGAGCGUAGAAACGCACUAAUUUCCAGUAGUGCAGCUCACACACAC------------AUGCAGA ......((((..(((((.((......(((((..((....))..)))))((((((....))((((((........)))))).)))))).)))))------------.)))).. ( -30.90) >consensus AAAAAAUGCGCAGUGUGGGUAAUUAAAAAAGUAGCAGCAGCCGCUUUUGAGCGUAGAAACGCACUAAUUUUCAGUAGUGCAGCUCACCCACACA__CACACACCCAUGCAGA ............((((((((......(((((..((....))..)))))((((((....))((((((........)))))).))))................))))))))... (-22.16 = -24.23 + 2.06)

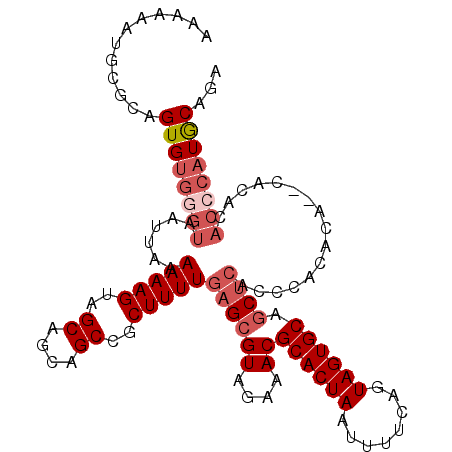
| Location | 3,115,955 – 3,116,057 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.09 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -21.38 |
| Energy contribution | -25.12 |
| Covariance contribution | 3.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3115955 102 - 27905053 UCUGCAUGGGUGGGUG--UGU--------GAUGCACUACUGAAAAUUAGUGCGUUUCUACGCUCAAAAGCGGCUGCUGCUACUUUUUUAAUUACCCACACUGCGCAUUUUUU ...(((((((((((((--((.--------(((((((((.(....).)))))))))..)))))))(((((..((....))..)))))......)))))...)))......... ( -35.80) >DroSec_CAF1 3660 110 - 1 UCUGCAUGUGUGUGUG--UGUAUGGGUGAGCUGCACUACUGAAAAUUAGUGCGUUUCUACGCUCAAAAGCGGCUGCUGCUACUUUUUUAAUUACCCACACUGCGCAUUUUUU .......(((((((.(--(((..((((((((.((((((.(....).))))))((....))))))(((((..((....))..)))))......)))))))))))))))..... ( -35.90) >DroSim_CAF1 8982 112 - 1 UCUGUAGGGGUGUGUGUAUUUGUGGGUGAGCUGCACUACUGAAAAUUAGUGCGUUUCUACGCUCAAAAGCGGCUGCUGCUACUUUUUUAAUUACCCACACUGCGCAUUUUUU ...........(((((((..(((((((((((.((((((.(....).))))))((....))))))(((((..((....))..)))))......))))))).)))))))..... ( -38.40) >DroYak_CAF1 7181 100 - 1 UCUGCAU------------GUGUGUGUGAGCUGCACUACUGGAAAUUAGUGCGUUUCUACGCUCAAAAGCGGCUGCUGCUACUUUUUUAAUUACCCACACUUCGCAUUUUUU ..(((..------------(((((.((((((.((((((.(....).))))))((....))))))(((((..((....))..)))))......)).)))))...)))...... ( -30.60) >consensus UCUGCAUGGGUGUGUG__UGUGUGGGUGAGCUGCACUACUGAAAAUUAGUGCGUUUCUACGCUCAAAAGCGGCUGCUGCUACUUUUUUAAUUACCCACACUGCGCAUUUUUU .............(((...((((((((((((.((((((.(....).))))))((....))))))(((((..((....))..)))))......))))))))..)))....... (-21.38 = -25.12 + 3.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:35 2006