
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,735,833 – 24,735,950 |
| Length | 117 |
| Max. P | 0.825928 |

| Location | 24,735,833 – 24,735,950 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -16.94 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
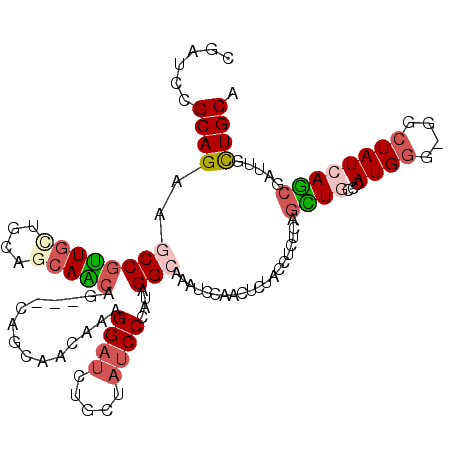
>3R_DroMel_CAF1 24735833 117 + 27905053 CGAUCCGCAGAAGCCGUUGCUGCAGCAACAGCAGCAGCAACAAAGGAUCUGCUAUCCCAUAGGCAAAUCUAACUCUACCUCUCAGCUACCCAUGGGCGGCUAUCAGCGAUUGCUGCA .(((((((....)).((((((((.((....)).))))))))...)))))((((........))))..................(((..((....))..)))..((((....)))).. ( -39.50) >DroVir_CAF1 20994 111 + 1 CGAUCCGCAGAAGCCGCUAAUGCAGCAGCAA---CAGCAACAAAGGAUGGCCUAUCCCAUAGGCAAAUCCAACUCGACCUCUCAGUUGCCCAUGG---GCUAUCAACGUCUACUGCA ......((((..((.(((.....))).))..---..(((((...((((.((((((...))))))..)))).....((....)).)))))...(((---((.......))))))))). ( -31.30) >DroPse_CAF1 10922 114 + 1 CGACCCGCAGAAGCCGUUGAUGCAACAACAGCAGCAGCAGC---GGAACUGCUAUCCCAUUGGAAAGUCGAAUUCUACCUCACAGCUACCGAUGGGUGGCUAUCAGCGAUUGUUGCA ((((..((((...((((((.(((..........))).))))---))..))))..(((....)))..)))).............(((((((....)))))))....((((...)))). ( -36.60) >DroGri_CAF1 9572 108 + 1 CGAUCCGCAGAAGCCGCUGUUGCAGCAGCA------GCAGCAGAGGAUGGCAUAUCCAAUUGGCAAGUCCAAUUCUACCUCUCAGCUGCCCAUGG---GCUAUCAGCGACUGCUGCA ......((((.((.(((((..((..((...------((((((((((.(((.....)))(((((.....)))))....)))))..)))))...)).---))...))))).)).)))). ( -40.10) >DroSec_CAF1 10400 117 + 1 CGAUCCGCAGAAGCCGUUGCUGCAGCAACAGCAGAAGCAACAAAGGCUCUGCUAUCCCAUAGGCAAAUCUAACUCUACCUCUCAGCUGCCCAUGGGCGGCUAUCAGCGAUUGCUGCA ......((((..((((((((....)))))((((((.((.......))))))))........)))...................(((((((....)))))))...........)))). ( -42.20) >DroWil_CAF1 8866 108 + 1 CGAUCCGCAGAAACCAUUGCUGCAGCAGCAG---------GCACGGAUUUGUUAUCCCAUAGGCAAGUCAAAUUCUACCUCACAAUUGCCCAUGGCUGGAUAUCAGCGAUUGUUGCA ......((((.........)))).(((((((---------.(..((((.....))))(((.(((((...................))))).)))((((.....))))).))))))). ( -28.91) >consensus CGAUCCGCAGAAGCCGUUGCUGCAGCAACAG___CAGCAACAAAGGAUCUGCUAUCCCAUAGGCAAAUCCAACUCUACCUCUCAGCUGCCCAUGGG_GGCUAUCAGCGAUUGCUGCA ......((((..((((((((....)))))...............((((.....))))....)))....................((((...((((....)))))))).....)))). (-16.94 = -17.80 + 0.86)

| Location | 24,735,833 – 24,735,950 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -43.07 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.17 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
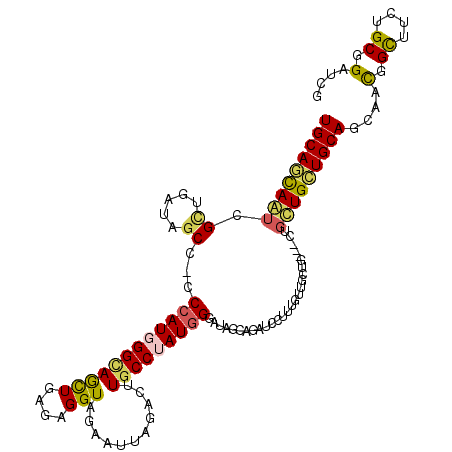
>3R_DroMel_CAF1 24735833 117 - 27905053 UGCAGCAAUCGCUGAUAGCCGCCCAUGGGUAGCUGAGAGGUAGAGUUAGAUUUGCCUAUGGGAUAGCAGAUCCUUUGUUGCUGCUGCUGUUGCUGCAGCAACGGCUUCUGCGGAUCG ..((((....))))...((..(((((((((((((....)))...........))))))))))...)).(((((..(((((((((.((....)).)))))))))((....))))))). ( -51.90) >DroVir_CAF1 20994 111 - 1 UGCAGUAGACGUUGAUAGC---CCAUGGGCAACUGAGAGGUCGAGUUGGAUUUGCCUAUGGGAUAGGCCAUCCUUUGUUGCUG---UUGCUGCUGCAUUAGCGGCUUCUGCGGAUCG ....(((((.(((.....(---((((((((((((....))((......)).)))))))))))...)))........(((((((---.(((....))).))))))).)))))...... ( -38.10) >DroPse_CAF1 10922 114 - 1 UGCAACAAUCGCUGAUAGCCACCCAUCGGUAGCUGUGAGGUAGAAUUCGACUUUCCAAUGGGAUAGCAGUUCC---GCUGCUGCUGCUGUUGUUGCAUCAACGGCUUCUGCGGGUCG (((((((((..((.(((((.(((....))).))))).))((((..........(((....)))((((((....---.)))))))))).)))))))))....(((((......))))) ( -39.10) >DroGri_CAF1 9572 108 - 1 UGCAGCAGUCGCUGAUAGC---CCAUGGGCAGCUGAGAGGUAGAAUUGGACUUGCCAAUUGGAUAUGCCAUCCUCUGCUGC------UGCUGCUGCAACAGCGGCUUCUGCGGAUCG .((((.((((((((.((((---.....((((((.((((((...((((((.....))))))((.....))..))))).).))------))))))))...)))))))).))))...... ( -45.50) >DroSec_CAF1 10400 117 - 1 UGCAGCAAUCGCUGAUAGCCGCCCAUGGGCAGCUGAGAGGUAGAGUUAGAUUUGCCUAUGGGAUAGCAGAGCCUUUGUUGCUUCUGCUGUUGCUGCAGCAACGGCUUCUGCGGAUCG ..((((....))))....((.(((((((((((((....)))...........))))))))))...((((((((.(((((((....((....)).))))))).))).))))))).... ( -51.40) >DroWil_CAF1 8866 108 - 1 UGCAACAAUCGCUGAUAUCCAGCCAUGGGCAAUUGUGAGGUAGAAUUUGACUUGCCUAUGGGAUAACAAAUCCGUGC---------CUGCUGCUGCAGCAAUGGUUUCUGCGGAUCG .......(((((((.....))))(((((((((..((.((((...)))).))))))))))).))).....((((((((---------((((((...)))))..)))....)))))).. ( -32.40) >consensus UGCAGCAAUCGCUGAUAGCC_CCCAUGGGCAGCUGAGAGGUAGAAUUAGACUUGCCUAUGGGAUAGCAGAUCCUUUGUUGCUG___CUGCUGCUGCAGCAACGGCUUCUGCGGAUCG (((((((((.((.....))...((((((((((((....)))...........)))))))))...........................)))))))))....(.((....)).).... (-24.38 = -24.17 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:30:35 2006