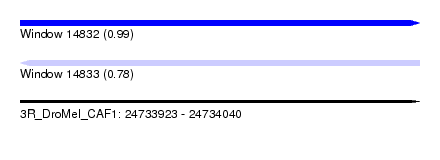
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,733,923 – 24,734,040 |
| Length | 117 |
| Max. P | 0.991115 |

| Location | 24,733,923 – 24,734,040 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.39 |
| Mean single sequence MFE | -43.03 |
| Consensus MFE | -14.42 |
| Energy contribution | -15.15 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.34 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
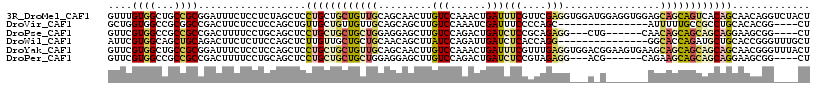
>3R_DroMel_CAF1 24733923 117 + 27905053 AGUAGACCUGUUGCUGUGACUGCUGCUCCACCUCCAUCCACCUCGAACGAAAUCAGUUUGGACAAGUUGCUGCAACAGCAGCAGGAGCUAGAGGAGAAAUCCGCGGCAGCCACAAAC (((((.....)))))(((.(((((((((...((((.......((((((.......))))))......((((((....))))))))))...))(((....)))))))))).))).... ( -42.60) >DroVir_CAF1 19067 98 + 1 AG----CCGUGUGCAGGCGGCAAAAAU---------------GCUGGGGAAAUCGAUUUGGACAAGCUGCUGCAACAACAGCAACAGCUGGAGGAGAAGUCGGCCGCGGCCACCAGC .(----((((......)))))......---------------(((((.......(((((...(.(((((.(((.......))).)))))...)...)))))(((....))).))))) ( -35.60) >DroPse_CAF1 8911 104 + 1 AG----CCGCUUCCUGCUGCUGCUGUUG------CAG---CCUCUGCGGAGAUCAGUCUGGACAAGCUCCUCCAGCAGCAGCAGGAGCUGCAGGAAAAGUCGGCGGCGGCCACGAAC .(----(((((((((((((((((((..(------(((---...))))((((....((....))...))))..))))))))))))))(((((.......).))))))))))....... ( -53.50) >DroWil_CAF1 6809 102 + 1 AGCAAACCCGGUGCAGCAUCUGGUGCC---------------CCUGGUGAGAUCAAUCUGGAUAAGCUGUUGCAGCAGCAACAAGAGCUGGAAGAGAAGUCUGCAGCUGCCACGAAU .....(((.((.(((.(....).))).---------------)).)))........(((((....(((((....)))))......(((((..(((....))).))))).))).)).. ( -29.30) >DroYak_CAF1 7384 117 + 1 AGUAAACCCGUUGCUGCUGCUGCUGCUUCACUUCCGUCCACCUCAAACGAAAUCAGUUUGGACAAGUUGCUGCAACAGCAGCAGGAGCUGGAGGAGAAAUCCGCGGCAGCCACGAAC ........(((.((((((((......((((.(((((((((.....(((.......))))))))....((((((....)))))))))).))))(((....))))))))))).)))... ( -47.90) >DroPer_CAF1 8975 104 + 1 AG----CCGCUUCCUGCUGCUGCUUCUG------CGU---CCUCUACGGAGAUCAGUCUGGACAAGCUCCUCCAGCAGCAGCAGGAGCUGCAGGAAAAGUCGGCGGCGGCCACGAAC .(----(((((((((((((((((..(((------.((---(.((....))))))))..((((........))))))))))))))))(((((.......).))))))))))....... ( -49.30) >consensus AG____CCGGUUGCUGCUGCUGCUGCU_______C_U___CCUCUAAGGAAAUCAGUCUGGACAAGCUGCUGCAACAGCAGCAGGAGCUGGAGGAGAAGUCGGCGGCGGCCACGAAC ...............((((((((.........................(....)..(((.....((((.((((.......)))).))))...))).......))))))))....... (-14.42 = -15.15 + 0.73)

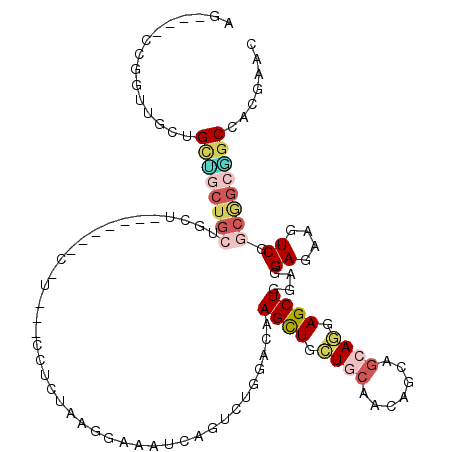
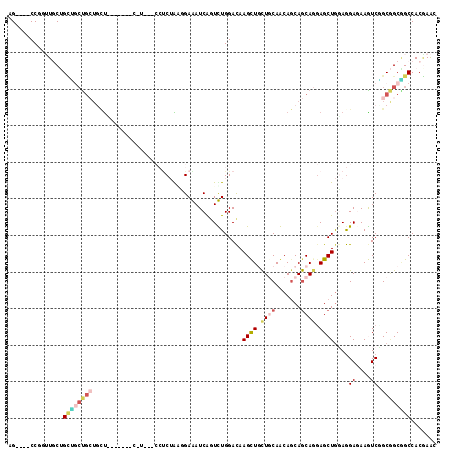
| Location | 24,733,923 – 24,734,040 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.39 |
| Mean single sequence MFE | -42.85 |
| Consensus MFE | -12.18 |
| Energy contribution | -13.38 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24733923 117 - 27905053 GUUUGUGGCUGCCGCGGAUUUCUCCUCUAGCUCCUGCUGCUGUUGCAGCAACUUGUCCAAACUGAUUUCGUUCGAGGUGGAUGGAGGUGGAGCAGCAGUCACAGCAACAGGUCUACU (((.((((((((.(((((....)))((((.((((((((((....))))))....(((((...(((......)))...))))))))).)))))).)))))))))))............ ( -49.20) >DroVir_CAF1 19067 98 - 1 GCUGGUGGCCGCGGCCGACUUCUCCUCCAGCUGUUGCUGUUGUUGCAGCAGCUUGUCCAAAUCGAUUUCCCCAGC---------------AUUUUUGCCGCCUGCACACGG----CU (((((.(((....)))(((.........(((((((((.......))))))))).))).............)))))---------------......((((........)))----). ( -35.00) >DroPse_CAF1 8911 104 - 1 GUUCGUGGCCGCCGCCGACUUUUCCUGCAGCUCCUGCUGCUGCUGGAGGAGCUUGUCCAGACUGAUCUCCGCAGAGG---CUG------CAACAGCAGCAGCAGGAAGCGG----CU ......((((((.((.(.......).))...((((((((((((((..((((...((....))....))))((((...---)))------)..)))))))))))))).))))----)) ( -51.00) >DroWil_CAF1 6809 102 - 1 AUUCGUGGCAGCUGCAGACUUCUCUUCCAGCUCUUGUUGCUGCUGCAACAGCUUAUCCAGAUUGAUCUCACCAGG---------------GGCACCAGAUGCUGCACCGGGUUUGCU ....(..(((((.(((((((........)).)).))).)))))..)...(((..((((.((......))....((---------------.(((.(....).))).))))))..))) ( -29.80) >DroYak_CAF1 7384 117 - 1 GUUCGUGGCUGCCGCGGAUUUCUCCUCCAGCUCCUGCUGCUGUUGCAGCAACUUGUCCAAACUGAUUUCGUUUGAGGUGGACGGAAGUGAAGCAGCAGCAGCAGCAACGGGUUUACU (..((((((((..(.(((....))).)))))).(((((((((((((..((..(((((((..((.(.......).)).)))))))...))..)))))))))))))..)))..)..... ( -45.20) >DroPer_CAF1 8975 104 - 1 GUUCGUGGCCGCCGCCGACUUUUCCUGCAGCUCCUGCUGCUGCUGGAGGAGCUUGUCCAGACUGAUCUCCGUAGAGG---ACG------CAGAAGCAGCAGCAGGAAGCGG----CU ......((((((.((.(.......).))...((((((((((((((((.(....).)))...(((...(((.....))---)..------))).))))))))))))).))))----)) ( -46.90) >consensus GUUCGUGGCCGCCGCCGACUUCUCCUCCAGCUCCUGCUGCUGCUGCAGCAGCUUGUCCAAACUGAUCUCCCCAGAGG___A_G_______AGCAGCAGCAGCAGCAACCGG____CU ....((((...))))..................((((((((((((.........(((......)))(((....)))................))))))))))))............. (-12.18 = -13.38 + 1.20)
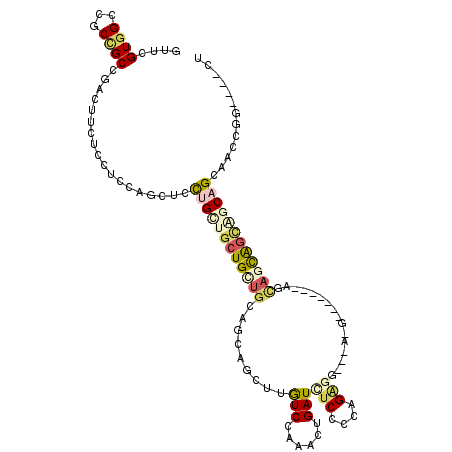
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:30:31 2006