
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,678,306 – 24,678,565 |
| Length | 259 |
| Max. P | 0.992770 |

| Location | 24,678,306 – 24,678,420 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -35.14 |
| Consensus MFE | -27.66 |
| Energy contribution | -27.82 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24678306 114 - 27905053 UGGCCCGGCAACAAUAAAUACUUCAACUAUAAAUAGGCAAAUGGCUACGAAAGGAAAAUA----CAUACAUUCGAAUGUGUGUAUUUUUAUGGAGCCUUAGUGUGUAUAUGGGGGUGU-- ..(((((....)..............(((((.(((.((.((.((((.(....)(((((((----(((((((....))))))))))))))....)))))).)).))).)))))))))..-- ( -37.10) >DroSec_CAF1 19222 114 - 1 UGGCCCGGCAACAAUAAAUACUUCAACUAUAAAUAGGCAAAUGGCUGCGAAAGGAAAAUA----CAGGCAUCCGAAUGUGUGUAUUUUUAUGGAGCCUUAGUGUGUAUAUGGGGGUGU-- ..(((((....)..............(((((.(((.((.((.((((.((....(((((((----((.((((....)))).))))))))).)).)))))).)).))).)))))))))..-- ( -36.10) >DroSim_CAF1 19306 114 - 1 UGGCCCGGCAACAAUAAAUACUUCAACUAUAAAUAGGCAAAUGGCUGCGAAAGGAAAAUA----CAUGCAUCCGAAUGUGAGUAUUUUUAUGGAGCCUUAGUGUGUAUAUGGGGGUGU-- ..(((((....)..............(((((.(((.((.((.((((.((....(((((((----(.(((((....))))).)))))))).)).)))))).)).))).)))))))))..-- ( -32.20) >DroEre_CAF1 20733 114 - 1 UGGCCCGGCAACAAUAAAUACUUCAACUAUAAAUAGGCAAAUGGCUGCAAAAGGAAAAUA----UAUGCAUCCGAAUGUGUGUAUUUUUAUGGAGCCUCUGUGUGUAUAUGGGGGUGU-- ..(((((....)..............(((((.(((.(((...((((.((....(((((((----(((((((....)))))))))))))).)).))))..))).))).)))))))))..-- ( -37.30) >DroYak_CAF1 19816 120 - 1 UGGCCCGGCAACAAUAAAUACUUCAACUAUAAAUAGGCAAAUGGCAGCAAAAGGAAAAUACAUGUAUGCAACCGAAUGUGUGUAUUUUUAUGGAGCCUUAGUGUGUAUAUGGGGGUGCCU .((((((....)..............(((((.(((.((.((.(((..((....(((((((((..(((........)))..))))))))).))..))))).)).))).))))).)).))). ( -33.00) >consensus UGGCCCGGCAACAAUAAAUACUUCAACUAUAAAUAGGCAAAUGGCUGCGAAAGGAAAAUA____CAUGCAUCCGAAUGUGUGUAUUUUUAUGGAGCCUUAGUGUGUAUAUGGGGGUGU__ ..(((((....)..............(((((.(((.((....((((.((....(((((((....(((((((....)))))))))))))).)).))))...)).))).))))))))).... (-27.66 = -27.82 + 0.16)



| Location | 24,678,420 – 24,678,539 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -43.92 |
| Consensus MFE | -40.02 |
| Energy contribution | -39.90 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24678420 119 + 27905053 CUCGUGGCAUGCUCAAGUGGCCACUCGAGGG-GGGCACUUUAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCGAGAAGUGGUAACGCAACUGGUUGUAAUCCAGCCGCUUCUU ...((((((.....(((((.((.((....))-)).)))))....))))))(((((((.....))(((...)))))))).((((((((((...((((....))))......)))))))))) ( -43.10) >DroSec_CAF1 19336 120 + 1 CUCGUGGCAUGCUCAAGUGGCCACUCGAGGGGGGGCACUUUAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCAAGAAGUGGCAACGCAACUGGUUGUAAUCCAGCCGCUUCUU ...((((((.....(((((.((.((....)).)).)))))....))))))(((((((.....))(((...)))))))).((((((((((...((((....))))......)))))))))) ( -44.40) >DroSim_CAF1 19420 118 + 1 CUCGUGGCAUGCUCAAGUGGCCACUCGAGG--GGGCACUUUAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCAAGAAGUGGCAACGCAACUGGUUGUAAUCCAGCCGCUUCUU ...((((((.....(((((.((........--)).)))))....))))))(((((((.....))(((...)))))))).((((((((((...((((....))))......)))))))))) ( -42.40) >DroEre_CAF1 20847 118 + 1 CUCGUGGCAUGCUCAAGUGGACACUCGUGG--GGGCACUUUAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCAAGAAGUGGCAACGCAACUGGUUGUAAUCCAGCCGCUUCUU .(((((((((((((..((((((...(((((--.((........)).)))))..))))))....)))).)))))))))..((((((((((...((((....))))......)))))))))) ( -47.10) >DroYak_CAF1 19936 118 + 1 CUCGUGGCAUGCUCAAGUGGCCACUCGUGG--GGGCACUUUAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCAAGAAGUGGCAACGCAACUGGUUGUAAUCCAGCCGCUUCUU ...((((((.....(((((.((.(....).--)).)))))....))))))(((((((.....))(((...)))))))).((((((((((...((((....))))......)))))))))) ( -42.60) >consensus CUCGUGGCAUGCUCAAGUGGCCACUCGAGG__GGGCACUUUAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCAAGAAGUGGCAACGCAACUGGUUGUAAUCCAGCCGCUUCUU ...((((((.....(((((.((.(....)...)).)))))....))))))(((((((.....))(((...)))))))).((((((((((...((((....))))......)))))))))) (-40.02 = -39.90 + -0.12)



| Location | 24,678,459 – 24,678,565 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 95.94 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -29.88 |
| Energy contribution | -29.76 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24678459 106 + 27905053 UAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCGAGAAGUGGUAACGCAACUGGUUGUAAUCCAGCCGCUUCUUGGACU------GUAGCUUUUCCUGCUCUCAAU ..((((((...(((.......)))))))))(..((((((((((((((((...((((....))))......)))))))))))).))------))..)................ ( -31.50) >DroSec_CAF1 19376 106 + 1 UAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCAAGAAGUGGCAACGCAACUGGUUGUAAUCCAGCCGCUUCUUAGACU------GUAGCUUUUCCUGCUCUCAUU .....((((.((.((((......))))...))..)))).((((((((((...((((....))))......)))))))))).((..------((((......))))..))... ( -29.60) >DroSim_CAF1 19458 106 + 1 UAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCAAGAAGUGGCAACGCAACUGGUUGUAAUCCAGCCGCUUCUUGGACU------GUAGCUUUUCCUGCUCUCAUU ..((((((...(((.......)))))))))(..((((((((((((((((...((((....))))......)))))))))))).))------))..)................ ( -33.40) >DroEre_CAF1 20885 106 + 1 UAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCAAGAAGUGGCAACGCAACUGGUUGUAAUCCAGCCGCUUCUUGGACU------GUAGCUUUUCCUGCUCUCAUU ..((((((...(((.......)))))))))(..((((((((((((((((...((((....))))......)))))))))))).))------))..)................ ( -33.40) >DroYak_CAF1 19974 112 + 1 UAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCAAGAAGUGGCAACGCAACUGGUUGUAAUCCAGCCGCUUCUUGGACUUCAGCUUUAGCUUUUCCUGCUCUCAUU ..((((((...(((.......)))))))))((...((((((((((((((...((((....))))......)))))))))))).))...))...(((.......)))...... ( -32.50) >consensus UAAUUGCCACGCCGUCCAUCCUGGGGCAAUGUCAUGGCCAAGAAGUGGCAACGCAACUGGUUGUAAUCCAGCCGCUUCUUGGACU______GUAGCUUUUCCUGCUCUCAUU ..((((((...(((.......))))))))).....((((((((((((((...((((....))))......)))))))))))).))........(((.......)))...... (-29.88 = -29.76 + -0.12)


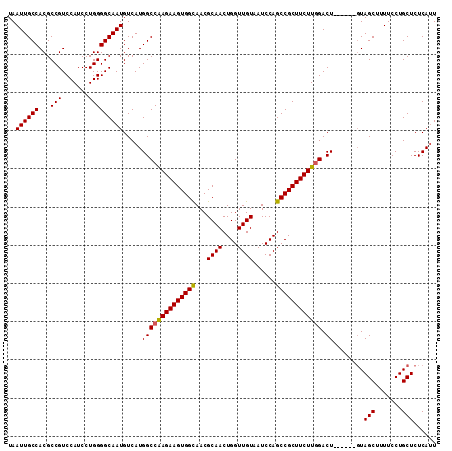
| Location | 24,678,459 – 24,678,565 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.94 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -29.94 |
| Energy contribution | -30.34 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
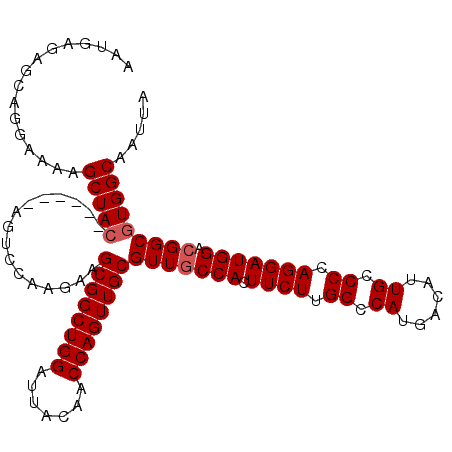
>3R_DroMel_CAF1 24678459 106 - 27905053 AUUGAGAGCAGGAAAAGCUAC------AGUCCAAGAAGCGGCUGGAUUACAACCAGUUGCGUUACCACUUCUCGGCCAUGACAUUGCCCCAGGAUGGACGGCGUGGCAAUUA ................(((((------.(((((....((((((((.......))))))))........((((.((.((......)).)).)))))))))...)))))..... ( -32.10) >DroSec_CAF1 19376 106 - 1 AAUGAGAGCAGGAAAAGCUAC------AGUCUAAGAAGCGGCUGGAUUACAACCAGUUGCGUUGCCACUUCUUGGCCAUGACAUUGCCCCAGGAUGGACGGCGUGGCAAUUA ................(((((------..........((((((((.......))))))))(((((((..((((((.((......))..))))))))).)))))))))..... ( -33.80) >DroSim_CAF1 19458 106 - 1 AAUGAGAGCAGGAAAAGCUAC------AGUCCAAGAAGCGGCUGGAUUACAACCAGUUGCGUUGCCACUUCUUGGCCAUGACAUUGCCCCAGGAUGGACGGCGUGGCAAUUA ................(((((------..........((((((((.......))))))))(((((((..((((((.((......))..))))))))).)))))))))..... ( -33.80) >DroEre_CAF1 20885 106 - 1 AAUGAGAGCAGGAAAAGCUAC------AGUCCAAGAAGCGGCUGGAUUACAACCAGUUGCGUUGCCACUUCUUGGCCAUGACAUUGCCCCAGGAUGGACGGCGUGGCAAUUA ................(((((------..........((((((((.......))))))))(((((((..((((((.((......))..))))))))).)))))))))..... ( -33.80) >DroYak_CAF1 19974 112 - 1 AAUGAGAGCAGGAAAAGCUAAAGCUGAAGUCCAAGAAGCGGCUGGAUUACAACCAGUUGCGUUGCCACUUCUUGGCCAUGACAUUGCCCCAGGAUGGACGGCGUGGCAAUUA ..........(((..(((....)))....))).....((((((((.......))))))))((((((((.((........))....(((((.....))..))))))))))).. ( -35.70) >consensus AAUGAGAGCAGGAAAAGCUAC______AGUCCAAGAAGCGGCUGGAUUACAACCAGUUGCGUUGCCACUUCUUGGCCAUGACAUUGCCCCAGGAUGGACGGCGUGGCAAUUA ................(((((................((((((((.......))))))))(((((((.((((.((.((......)).)).))))))).)))))))))..... (-29.94 = -30.34 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:30:12 2006