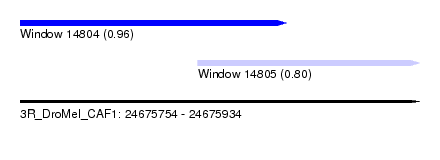
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,675,754 – 24,675,934 |
| Length | 180 |
| Max. P | 0.957108 |

| Location | 24,675,754 – 24,675,874 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -31.70 |
| Energy contribution | -32.38 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24675754 120 + 27905053 UAGGAGCCAAUUUUAGUGGUUUAGUCCAGCCACUUGACCCACAUUCCGGUGGUUUCCCUUUCCGCUGCCGGCGAGCGAGUUCCACUUUGUCGGCGACCUCAUAUUGUUGGCUAUAACACA ....(((((((..((((((...((...(((((((.((.......)).)))))))...))..))))((((((((((.(.....).).)))))))))......))..)))))))........ ( -37.30) >DroSec_CAF1 16772 120 + 1 UAGGAGCUAAUUUUAGUGGUUUAGGCCAGCCACUUGACCCACAUUCCGGUGGUUUCCCCUUCCGCAGCCGGCGAGCGAGUUCCACUUUGUCGGCGACCUCAUAUUGCUGGCUAUAACACA ..((((((.......((((...(((..(((((((.((.......)).)))))))...))).)))).((......)).)))))).....((((((((.......))))))))......... ( -36.60) >DroSim_CAF1 16822 120 + 1 UAGGAGCUAAUUUUAGUGGUUUAGGCCAGCCACUUGACCCACAUUCCGGUGGUUUCCCUUUCCGCAGUCGGCGAGCGAGUUCCACUUUGUCGGCGACCUCAUAUUGCUGGCUAUAACACA ..(((((........((((...(((..(((((((.((.......)).)))))))..)))..))))..(((.....)))))))).....((((((((.......))))))))......... ( -37.00) >DroEre_CAF1 18195 114 + 1 --GUAGCUAAUUUUAGUGGUUUAGGCCAGCCACUUGACCCACAUUCCCGUGGUUUCCCUUUCCGCAGCCGGC----GAGUUCCACUUUGUCGGCGACCUCAUAUUGCUGGCUAUAACACA --(((((((......((((...(((..((((((..((.......))..))))))..)))..)))).((((((----(((......))))))))).............)))))))...... ( -37.50) >DroYak_CAF1 17242 116 + 1 UCGUAGCUAAUUUUAGUGGUUUUGGCCAGCCACUUGACCCACAUUCCCGUGGUUUCCCUUUCCGCAGCCGGC----GAGUUCCACUUUGUCGGCGACCUCAUAUUGCUGGCUAUAACACA ...............(((....((((((((....(((.((((......))))..............((((((----(((......)))))))))....)))....))))))))...))). ( -37.50) >consensus UAGGAGCUAAUUUUAGUGGUUUAGGCCAGCCACUUGACCCACAUUCCGGUGGUUUCCCUUUCCGCAGCCGGCGAGCGAGUUCCACUUUGUCGGCGACCUCAUAUUGCUGGCUAUAACACA ....(((((......((((...(((..(((((((.((.......)).)))))))..)))..)))).((((((....(((.....))).)))))).............)))))........ (-31.70 = -32.38 + 0.68)


| Location | 24,675,834 – 24,675,934 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -33.99 |
| Consensus MFE | -28.65 |
| Energy contribution | -28.53 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24675834 100 + 27905053 UCCACUUUGUCGGCGACCUCAUAUUGUUGGCUAUAACACAAUUAAUUAAUUAAAUUCUUAAGCCUCCCCAGUGGCACUCGAGUGCAGGGGCGUCGCAGGG-------------------- ....(((((.(((((.((((.(((((..((((............................))))....)))))((((....)))).))))))))))))))-------------------- ( -28.59) >DroSec_CAF1 16852 105 + 1 UCCACUUUGUCGGCGACCUCAUAUUGCUGGCUAUAACACAAUUAAUUAAUUAAAUUCUUAAGCCUCCCCAGUGGCACACGAGUGCAGAGGCGUUGCAGGG-GG--------------GAG (((.(((((.(((((.((((.(((((..((((............................))))....)))))((((....)))).))))))))))))))-.)--------------)). ( -32.39) >DroSim_CAF1 16902 106 + 1 UCCACUUUGUCGGCGACCUCAUAUUGCUGGCUAUAACACAAUUAAUUAAUUAAAUUCUUAAGCCUCCCCAGUGGCACACGAGUGCAGGGGCGUCGCAGGGGGG--------------GAG (((.(((((.(((((.((((.(((((..((((............................))))....)))))((((....)))).))))))))))))))..)--------------)). ( -33.49) >DroEre_CAF1 18269 102 + 1 UCCACUUUGUCGGCGACCUCAUAUUGCUGGCUAUAACACAAUUAAUUAAUUAAAUUCUUAAGCCUCCCCAGUGGCACUUGAGUGCAGGGGCGUCGCUGGGGG------------------ ........((((((((.......))))))))..................................((((((((((.((((....))))...)))))))))).------------------ ( -31.30) >DroYak_CAF1 17318 120 + 1 UCCACUUUGUCGGCGACCUCAUAUUGCUGGCUAUAACACAAUUAAUUAAUUAAAUUCUUAAGCCUCCCCAGUGGCACUUGAGUGCAGGGGCGUCGCAGGGGGGGCAGAGGCGUCGUAGAA .....((((.(((((.((((..((((.((.......))))))...................((((((((.(((((.((((....))))...))))).)))))))).))))))))))))). ( -44.20) >consensus UCCACUUUGUCGGCGACCUCAUAUUGCUGGCUAUAACACAAUUAAUUAAUUAAAUUCUUAAGCCUCCCCAGUGGCACUCGAGUGCAGGGGCGUCGCAGGGGGG______________GA_ ....(((((.(((((.((((.(((((..((((............................))))....)))))((((....)))).)))))))))))))).................... (-28.65 = -28.53 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:30:07 2006