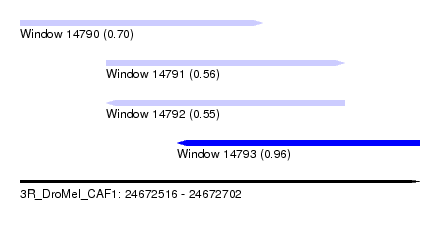
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,672,516 – 24,672,702 |
| Length | 186 |
| Max. P | 0.959214 |

| Location | 24,672,516 – 24,672,629 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24672516 113 + 27905053 UAAGCAGAAUAAAUAAACAGCAAAUAUAUUUUUGCCUGCACCACAGUCUACAUUUAAACGGUUACCACUG-------UGUAACCGGCGUGGAAAAUCAAGUUGCGUGUUGUGAUAACACA ...((((..........(((((((......)))).)))..((((.(....).......(((((((.....-------.)))))))..)))).........))))((((((...)))))). ( -26.70) >DroSec_CAF1 13554 113 + 1 UUAGCGGAAUAAAUAAACAGCAAAUAUAUUUUCCCCUGCACUACAGUCUACAUUUCAACGGUUACCACUG-------UGUAACAGGCGUGGAAAAUCAAGUUGCGUGUUGUGAUAACACA ...................((((....(((((((((((...((((((..((.........))....))))-------))...))))...)))))))....))))((((((...)))))). ( -24.10) >DroSim_CAF1 13548 113 + 1 UUAGCAGAAUAAAUAAACAGCAAAUAUAUUUUCCCCUGCACUACGGUCUACAUUUCAACGGUUACCACUG-------UGUAACCGGCGUGGAAAAUCAAGUUGCGUGUUGUGAUAACACA ...................((((....(((((((..(((((....))...........(((((((.....-------.)))))))))).)))))))....))))((((((...)))))). ( -28.60) >DroEre_CAF1 15060 113 + 1 UUAGCAAAAUAAAUAGACAGAAAAUAUAUUUUUCUCUGCACAAUAGUCGACAUUUCAACGGUUACCACUG-------UGUAACCGGCAUGGAAAAUCAAGUUGCGUGUUUUGAUAACACA ...((((........(((((((((.....))))))..........)))((..(((((.(((((((.....-------.)))))))...)))))..))...))))(((((.....))))). ( -25.70) >DroYak_CAF1 13850 120 + 1 UUAGCAAAAUAAAUAGACAGCAAGUAUAUUUUCCUCUGCACAAUAGUCGAGAUUUCAACGGUUACCAGUGUGUAGUGUGUAACCGGCGUGGAAAAUCAAGUUGCGUGUUUUGAUAACACA ...................((((.(..(((((((.(.((.........(......)..(((((((((.(....).)).)))))))))).)))))))..).))))(((((.....))))). ( -28.50) >consensus UUAGCAGAAUAAAUAAACAGCAAAUAUAUUUUCCCCUGCACAACAGUCUACAUUUCAACGGUUACCACUG_______UGUAACCGGCGUGGAAAAUCAAGUUGCGUGUUGUGAUAACACA ...................((((....(((((((..(((((....))...........(((((((.............)))))))))).)))))))....))))(((((.....))))). (-20.18 = -20.88 + 0.70)



| Location | 24,672,556 – 24,672,667 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.02 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24672556 111 + 27905053 CCACAGUCUACAUUUAAACGGUUACCACUG-------UGUAACCGGCGUGGAAAAUCAAGUUGCGUGUUGUGAUAACACACAUGCGAAUAAUACAAG--UGAAUCUUGGAUAUUCUUUGG ((((.(....).......(((((((.....-------.)))))))..)))).....((((..((((((.(((....)))))))))(((((...((((--.....))))..))))))))). ( -31.00) >DroSec_CAF1 13594 113 + 1 CUACAGUCUACAUUUCAACGGUUACCACUG-------UGUAACAGGCGUGGAAAAUCAAGUUGCGUGUUGUGAUAACACACAUGCGAACAAUACAAGUGUGGAUCUUGGGUAUUCUUUGG (((..((((((((((....((((.((((.(-------(.......))))))..))))..(((((((((.(((....))))))))).))).....))))))))))..)))........... ( -29.40) >DroSim_CAF1 13588 113 + 1 CUACGGUCUACAUUUCAACGGUUACCACUG-------UGUAACCGGCGUGGAAAAUCAAGUUGCGUGUUGUGAUAACACACAUGCGAACAAUACAAGAGUGGAUCUUGGGUAUUCUUUGG (((.(((((((.(((((.(((((((.....-------.)))))))...)))))......(((((((((.(((....))))))))).))).........))))))).)))........... ( -36.00) >DroEre_CAF1 15100 111 + 1 CAAUAGUCGACAUUUCAACGGUUACCACUG-------UGUAACCGGCAUGGAAAAUCAAGUUGCGUGUUUUGAUAACACACAUGCGAACAACACAAG--UGGAUCUUGGAUAUUCUUUGU ..................(((((((.....-------.)))))))(((.((((.(((..(((((((((..((....)).)))))).)))....((((--.....))))))).)))).))) ( -27.10) >DroYak_CAF1 13890 118 + 1 CAAUAGUCGAGAUUUCAACGGUUACCAGUGUGUAGUGUGUAACCGGCGUGGAAAAUCAAGUUGCGUGUUUUGAUAACACACAUGCGAACAAUACAAG--UGGAUCUUGGAUAUUCUUUGU .((((..((((((((...(((((((((.(....).)).)))))))((((((.....).......(((((.....))))).)))))............--.))))))))..))))...... ( -29.30) >consensus CAACAGUCUACAUUUCAACGGUUACCACUG_______UGUAACCGGCGUGGAAAAUCAAGUUGCGUGUUGUGAUAACACACAUGCGAACAAUACAAG__UGGAUCUUGGAUAUUCUUUGG ..................(((((((.............))))))).((.((((.(((...((((((((.(((....)))))))))))......((((.......))))))).)))).)). (-23.70 = -23.90 + 0.20)


| Location | 24,672,556 – 24,672,667 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.02 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24672556 111 - 27905053 CCAAAGAAUAUCCAAGAUUCA--CUUGUAUUAUUCGCAUGUGUGUUAUCACAACACGCAACUUGAUUUUCCACGCCGGUUACA-------CAGUGGUAACCGUUUAAAUGUAGACUGUGG .(((.(((((..((((.....--))))...)))))...((((((((.....))))))))..))).....((((...((((((.-------.....))))))(((((....))))).)))) ( -29.10) >DroSec_CAF1 13594 113 - 1 CCAAAGAAUACCCAAGAUCCACACUUGUAUUGUUCGCAUGUGUGUUAUCACAACACGCAACUUGAUUUUCCACGCCUGUUACA-------CAGUGGUAACCGUUGAAAUGUAGACUGUAG .....(((((..((((.......))))...)))))(((((((((((.....)))))))).((....((((.(((...(((((.-------.....)))))))).))))...))..))).. ( -22.40) >DroSim_CAF1 13588 113 - 1 CCAAAGAAUACCCAAGAUCCACUCUUGUAUUGUUCGCAUGUGUGUUAUCACAACACGCAACUUGAUUUUCCACGCCGGUUACA-------CAGUGGUAACCGUUGAAAUGUAGACCGUAG ............(((((.....)))))(((.(((..((((((((((.....)))))))..............((.(((((((.-------.....))))))).))..)))..))).))). ( -27.20) >DroEre_CAF1 15100 111 - 1 ACAAAGAAUAUCCAAGAUCCA--CUUGUGUUGUUCGCAUGUGUGUUAUCAAAACACGCAACUUGAUUUUCCAUGCCGGUUACA-------CAGUGGUAACCGUUGAAAUGUCGACUAUUG .....(((((.(((((.....--)))).).)))))...((((((((.....))))))))..(((((.((.((...(((((((.-------.....))))))).)).)).)))))...... ( -28.00) >DroYak_CAF1 13890 118 - 1 ACAAAGAAUAUCCAAGAUCCA--CUUGUAUUGUUCGCAUGUGUGUUAUCAAAACACGCAACUUGAUUUUCCACGCCGGUUACACACUACACACUGGUAACCGUUGAAAUCUCGACUAUUG .(((.(((((..((((.....--))))...)))))...((((((((.....))))))))..)))............((((((.((........))))))))(((((....)))))..... ( -27.00) >consensus CCAAAGAAUAUCCAAGAUCCA__CUUGUAUUGUUCGCAUGUGUGUUAUCACAACACGCAACUUGAUUUUCCACGCCGGUUACA_______CAGUGGUAACCGUUGAAAUGUAGACUGUAG .(((.(((((..((((.......))))...)))))...((((((((.....))))))))..)))...........(((((((.............))))))).................. (-20.98 = -21.02 + 0.04)


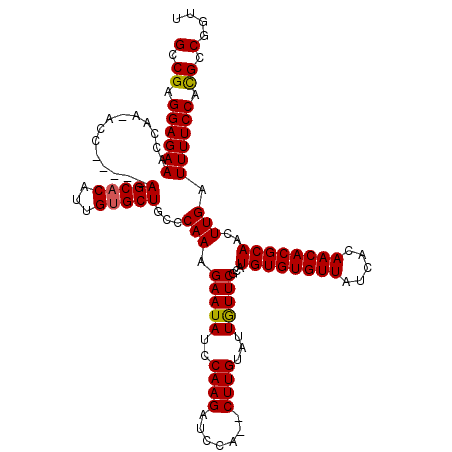
| Location | 24,672,589 – 24,672,702 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.79 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.22 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24672589 113 - 27905053 GUCGAGGAGAAACCAAGACC-----AGCACAUUGUGCUGCCCAAAGAAUAUCCAAGAUUCA--CUUGUAUUAUUCGCAUGUGUGUUAUCACAACACGCAACUUGAUUUUCCACGCCGGUU (.((.((((((..((((..(-----(((((...))))))......(((((..((((.....--))))...)))))...((((((((.....)))))))).)))).)))))).)).).... ( -32.00) >DroSec_CAF1 13627 115 - 1 GUCGAGGAGAAACCAAAACC-----AGCACAUUGUGCUGCCCAAAGAAUACCCAAGAUCCACACUUGUAUUGUUCGCAUGUGUGUUAUCACAACACGCAACUUGAUUUUCCACGCCUGUU (.((.((((((........(-----(((((...))))))..(((.(((((..((((.......))))...)))))...((((((((.....))))))))..))).)))))).)).).... ( -28.00) >DroSim_CAF1 13621 115 - 1 GACGAGGAGAAACCAAAACC-----AGCACAUUGUGCUGCCCAAAGAAUACCCAAGAUCCACUCUUGUAUUGUUCGCAUGUGUGUUAUCACAACACGCAACUUGAUUUUCCACGCCGGUU (.((.((((((........(-----(((((...))))))..(((.(((((..(((((.....)))))...)))))...((((((((.....))))))))..))).)))))).)).).... ( -30.40) >DroEre_CAF1 15133 111 - 1 GCCGAGGAGAAACCAA--CC-----AGCACAAUGUGCUGCACAAAGAAUAUCCAAGAUCCA--CUUGUGUUGUUCGCAUGUGUGUUAUCAAAACACGCAACUUGAUUUUCCAUGCCGGUU ((((.((((((.....--.(-----(((((...))))))..(((.(((((.(((((.....--)))).).)))))...((((((((.....))))))))..))).))))))....)))). ( -31.90) >DroYak_CAF1 13930 116 - 1 GCCGAGGAGAAACCAA--CUGGUGGAGCUCAAUGUGCUGCACAAAGAAUAUCCAAGAUCCA--CUUGUAUUGUUCGCAUGUGUGUUAUCAAAACACGCAACUUGAUUUUCCACGCCGGUU ((((.((((((.....--...(((.(((.......))).)))...(((((..((((.....--))))...)))))...((((((((.....))))))))......))))))....)))). ( -32.00) >consensus GCCGAGGAGAAACCAA_ACC_____AGCACAUUGUGCUGCCCAAAGAAUAUCCAAGAUCCA__CUUGUAUUGUUCGCAUGUGUGUUAUCACAACACGCAACUUGAUUUUCCACGCCGGUU (.((.((((((..............(((((...)))))...(((.(((((..((((.......))))...)))))...((((((((.....))))))))..))).)))))).)).).... (-25.34 = -25.22 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:29:57 2006