
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,671,780 – 24,671,977 |
| Length | 197 |
| Max. P | 0.999895 |

| Location | 24,671,780 – 24,671,900 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -45.04 |
| Consensus MFE | -38.60 |
| Energy contribution | -40.20 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24671780 120 + 27905053 CCCCGCUGGGCGUCACGUGCGCGGCCUUUGUUUUUAUUGCUGCCUUUUGCGGCGAUUCACGCGUUUUUCCGCGAAACGCCGGAUACCAUGCCAACCACCCCCGCACGUUACCGAGCGGGG (((((((.((....(((((((.(((.(((......((((((((.....))))))))...((((......))))))).)))((........)).........)))))))..)).))))))) ( -50.10) >DroSec_CAF1 12862 120 + 1 CCCCGCUGGGCGUCACGUGCGCGGCCUUUGUUUUUAUUGCUGCCUUUUGCGGCGAUUCACGCGUUUUUCCGCGAAACGCCGGAUACCAUGCCAACCACCCCCGCUCUUUUCCGAGCGGGG ....(((((..(((.(((.(((((....(((....((((((((.....))))))))....))).....)))))..)))...))).))).)).......((((((((......)))))))) ( -46.40) >DroSim_CAF1 12863 120 + 1 CCCCGCUGGGCGUCACGUGCGCGGCCUUUGUUUUUAUUGCUGCCUUUUGCGGCGAUUCACGCGUUUUUCCGCGAAACGCCGGAUACCAUGCCAACCACCCCCGCACUUUUCCGAGCGGGG (((((((.((......(((((.(((.(((......((((((((.....))))))))...((((......))))))).)))((........)).........)))))....)).))))))) ( -45.70) >DroEre_CAF1 14372 120 + 1 CCCCGCUGGGCGUCACGUGCGCGGCCUUUGUUUUUAUUGCUGCCUUUUGCGGCGAUUCACGCUUUUUUCCGCGAAACGCCGGAUGCUAUGGCAACCACCCCCGCACUUUUCCGUGCAGGG ....((((((((((.(((.(((((.....((....((((((((.....))))))))....))......)))))..)))...)))))).))))......(((.((((......)))).))) ( -43.20) >DroYak_CAF1 13061 120 + 1 CCCCGCUGGGCGUCACGUGCGCGGCCUUUGUUUUUAUUGCUGCCUUUUGCGGCGAUUCACGCUUUUUUCCGCGAAACGCCGGAUGCCAUGCCAACCACCCCCGCACUUUUCCGAGCAGGG (((.(((.((((((.(((.(((((.....((....((((((((.....))))))))....))......)))))..)))...)))))).(((...........)))........))).))) ( -39.80) >consensus CCCCGCUGGGCGUCACGUGCGCGGCCUUUGUUUUUAUUGCUGCCUUUUGCGGCGAUUCACGCGUUUUUCCGCGAAACGCCGGAUACCAUGCCAACCACCCCCGCACUUUUCCGAGCGGGG (((((((.(((((......(((((....(((....((((((((.....))))))))....))).....)))))..)))))(((.....(((...........)))....))).))))))) (-38.60 = -40.20 + 1.60)



| Location | 24,671,780 – 24,671,900 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -50.97 |
| Consensus MFE | -48.78 |
| Energy contribution | -48.70 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24671780 120 - 27905053 CCCCGCUCGGUAACGUGCGGGGGUGGUUGGCAUGGUAUCCGGCGUUUCGCGGAAAAACGCGUGAAUCGCCGCAAAAGGCAGCAAUAAAAACAAAGGCCGCGCACGUGACGCCCAGCGGGG (((((((.(((.(((((((.((.(.(((.((.........((((.((((((........)))))).))))((.....)).))......)))...).)).)))))))...))).))))))) ( -56.30) >DroSec_CAF1 12862 120 - 1 CCCCGCUCGGAAAAGAGCGGGGGUGGUUGGCAUGGUAUCCGGCGUUUCGCGGAAAAACGCGUGAAUCGCCGCAAAAGGCAGCAAUAAAAACAAAGGCCGCGCACGUGACGCCCAGCGGGG ((((((((......))))..(((((..((((.((((....((((.((((((........)))))).))))((........)).............)))).)).))...)))))...)))) ( -48.20) >DroSim_CAF1 12863 120 - 1 CCCCGCUCGGAAAAGUGCGGGGGUGGUUGGCAUGGUAUCCGGCGUUUCGCGGAAAAACGCGUGAAUCGCCGCAAAAGGCAGCAAUAAAAACAAAGGCCGCGCACGUGACGCCCAGCGGGG (((((((.((....(((((.((.(.(((.((.........((((.((((((........)))))).))))((.....)).))......)))...).)).)))))......)).))))))) ( -50.90) >DroEre_CAF1 14372 120 - 1 CCCUGCACGGAAAAGUGCGGGGGUGGUUGCCAUAGCAUCCGGCGUUUCGCGGAAAAAAGCGUGAAUCGCCGCAAAAGGCAGCAAUAAAAACAAAGGCCGCGCACGUGACGCCCAGCGGGG ((((((..((....(((((.((.(.((((((........(((((.((((((........)))))).))))).....))))))............).)).)))))......))..)))))) ( -48.74) >DroYak_CAF1 13061 120 - 1 CCCUGCUCGGAAAAGUGCGGGGGUGGUUGGCAUGGCAUCCGGCGUUUCGCGGAAAAAAGCGUGAAUCGCCGCAAAAGGCAGCAAUAAAAACAAAGGCCGCGCACGUGACGCCCAGCGGGG (((((((.((....(((((.((.(.(((......((....((((.((((((........)))))).))))((.....)).))......)))...).)).)))))......)).))))))) ( -50.70) >consensus CCCCGCUCGGAAAAGUGCGGGGGUGGUUGGCAUGGUAUCCGGCGUUUCGCGGAAAAACGCGUGAAUCGCCGCAAAAGGCAGCAAUAAAAACAAAGGCCGCGCACGUGACGCCCAGCGGGG (((((((.((....(((((.((.(.(((......((....((((.((((((........)))))).))))((.....)).))......)))...).)).)))))......)).))))))) (-48.78 = -48.70 + -0.08)

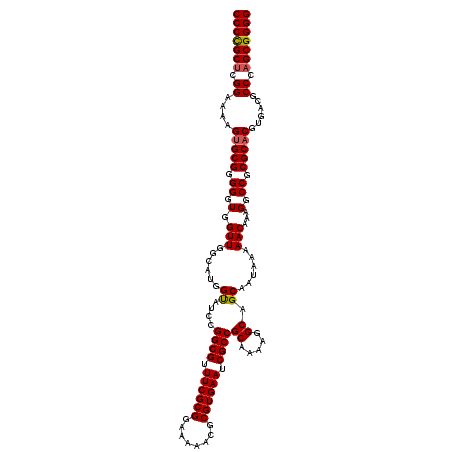

| Location | 24,671,820 – 24,671,940 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -38.74 |
| Energy contribution | -38.10 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.65 |
| SVM RNA-class probability | 0.999491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24671820 120 - 27905053 CAGUUUGUGUACAAAAAUAAGUAGAAAAGUCGCUUUUCCUCCCCGCUCGGUAACGUGCGGGGGUGGUUGGCAUGGUAUCCGGCGUUUCGCGGAAAAACGCGUGAAUCGCCGCAAAAGGCA ...((((....)))).............((((((...(((((((((.((....)).))))))).))..)))........(((((.((((((........)))))).))))).....))). ( -44.70) >DroSec_CAF1 12902 120 - 1 CAGUUUGUGUACAAAAAUAAGUAGAAAAAUCGCUUUUCCUCCCCGCUCGGAAAAGAGCGGGGGUGGUUGGCAUGGUAUCCGGCGUUUCGCGGAAAAACGCGUGAAUCGCCGCAAAAGGCA (..(((((((((...........((....))(((...(((((((((((......))))))))).))..)))...))))..((((.((((((........)))))).)))))))))..).. ( -45.20) >DroSim_CAF1 12903 120 - 1 CAGUUUGUGUACAAAAAUAAGUAGAAAAAUCGCUUUUCCUCCCCGCUCGGAAAAGUGCGGGGGUGGUUGGCAUGGUAUCCGGCGUUUCGCGGAAAAACGCGUGAAUCGCCGCAAAAGGCA (..(((((((((...........((....))(((...(((((((((.(......).))))))).))..)))...))))..((((.((((((........)))))).)))))))))..).. ( -41.00) >DroEre_CAF1 14412 120 - 1 CAGUUUUUGUACAAAAAUAAGUAGAAGAAUCGCUUUUCCUCCCUGCACGGAAAAGUGCGGGGGUGGUUGCCAUAGCAUCCGGCGUUUCGCGGAAAAAAGCGUGAAUCGCCGCAAAAGGCA ..((((((.(((........))).)))))).((((((...((((((((......))))))))((((.(((....))).))((((.((((((........)))))).)))))).)))))). ( -46.50) >DroYak_CAF1 13101 120 - 1 CAGUUUGUGUACGAAAAUAAGUAGAAGAAUCGCUUUUCCUCCCUGCUCGGAAAAGUGCGGGGGUGGUUGGCAUGGCAUCCGGCGUUUCGCGGAAAAAAGCGUGAAUCGCCGCAAAAGGCA ..((((.(.(((........))).).)))).(((...(((((((((.(......).))))))).))..)))...((...(((((.((((((........)))))).)))))......)). ( -40.10) >consensus CAGUUUGUGUACAAAAAUAAGUAGAAAAAUCGCUUUUCCUCCCCGCUCGGAAAAGUGCGGGGGUGGUUGGCAUGGUAUCCGGCGUUUCGCGGAAAAACGCGUGAAUCGCCGCAAAAGGCA ..((((.(.(((........))).).)))).(((((((((((((((.(......).))))))).)).............(((((.((((((........)))))).)))))..)))))). (-38.74 = -38.10 + -0.64)



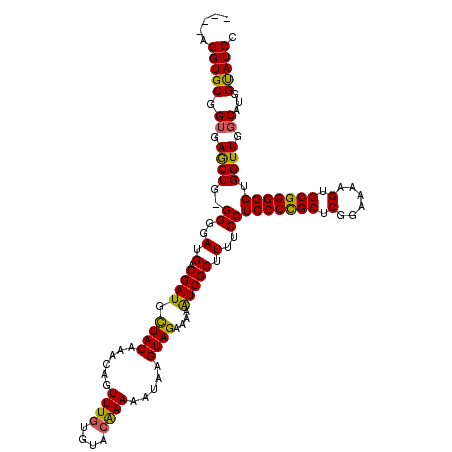
| Location | 24,671,860 – 24,671,977 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -39.24 |
| Consensus MFE | -34.02 |
| Energy contribution | -33.30 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24671860 117 - 27905053 --GGCGGUGCGGUGAACUG-GGGGAUGAUGAUGCUACAAACAGUUUGUGUACAAAAAUAAGUAGAAAAGUCGCUUUUCCUCCCCGCUCGGUAACGUGCGGGGGUGGUUGGCAUGGUAUCC --...(((((.(((.((((-(..((((((....((((......((((....)))).....))))....)))).))..))(((((((.((....)).))))))).)))...))).))))). ( -41.10) >DroSec_CAF1 12942 115 - 1 ----AGGUGCGGUGAGCUG-GGGGAUGAUGAUGCUACAAACAGUUUGUGUACAAAAAUAAGUAGAAAAAUCGCUUUUCCUCCCCGCUCGGAAAAGAGCGGGGGUGGUUGGCAUGGUAUCC ----.(((((.(((.((((-(..((((((....((((......((((....)))).....))))....)))).))..))(((((((((......))))))))).)))...))).))))). ( -42.80) >DroSim_CAF1 12943 115 - 1 ----AGGUGCGGUGAGCUG-GGGGAUGAUGAUGCUACAAACAGUUUGUGUACAAAAAUAAGUAGAAAAAUCGCUUUUCCUCCCCGCUCGGAAAAGUGCGGGGGUGGUUGGCAUGGUAUCC ----.(((((.(((.((((-(..((((((....((((......((((....)))).....))))....)))).))..))(((((((.(......).))))))).)))...))).))))). ( -38.60) >DroEre_CAF1 14452 119 - 1 AAGAAGGUGCGGUAAGCUG-GGGGAUGAUGAUGUUACAAACAGUUUUUGUACAAAAAUAAGUAGAAGAAUCGCUUUUCCUCCCUGCACGGAAAAGUGCGGGGGUGGUUGCCAUAGCAUCC .....((((((((((.(.(-(..((((((....((((.....((((((....))))))..))))....)))).))..))(((((((((......))))))))).).)))))...))))). ( -38.40) >DroYak_CAF1 13141 118 - 1 --AAAGGUGCGGUAAGCUGAGGGGAUGAUGAUGCUACAAACAGUUUGUGUACGAAAAUAAGUAGAAGAAUCGCUUUUCCUCCCUGCUCGGAAAAGUGCGGGGGUGGUUGGCAUGGCAUCC --...(((((.((..((..(((..(.(.((((.((((......((((....)))).....))))....))))).)..))(((((((.(......).)))))))...)..)))).))))). ( -35.30) >consensus ____AGGUGCGGUGAGCUG_GGGGAUGAUGAUGCUACAAACAGUUUGUGUACAAAAAUAAGUAGAAAAAUCGCUUUUCCUCCCCGCUCGGAAAAGUGCGGGGGUGGUUGGCAUGGUAUCC .....(((((.((.((((..((..(.(.((((.((((......((((....)))).....))))....))))).)..))(((((((.(......).))))))).)))).))...))))). (-34.02 = -33.30 + -0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:29:52 2006