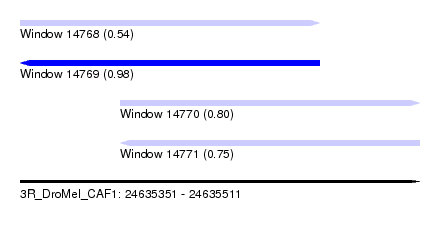
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,635,351 – 24,635,511 |
| Length | 160 |
| Max. P | 0.978565 |

| Location | 24,635,351 – 24,635,471 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.08 |
| Mean single sequence MFE | -40.38 |
| Consensus MFE | -32.14 |
| Energy contribution | -32.20 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
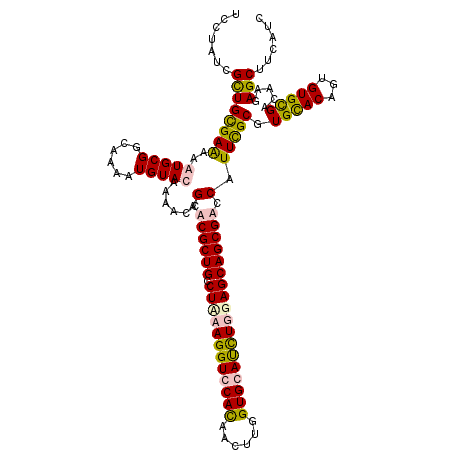
>3R_DroMel_CAF1 24635351 120 + 27905053 CUGUGUGUAUGUUGGAGUGCUGCUUAAGCUUCGAAUUGCUGAUGAAGCUCUUGUCGCACACUGUGCACGCGAAUGGUCGCUGCUCAAGAUGCACCAAGUUGUGGACCUUGAGACAGCGCU .(((((((((...((.((((.((...(((((((.........)))))))...)).)))).)))))))))))...((.(((((((((((.(.(((......))).).)))))).))))))) ( -45.40) >DroVir_CAF1 131767 120 + 1 CUGUAUGAAUGUUCGAGUGCUGUUUCAGCUUCGAGUUGCUAAUGAAGCUCUUGUCGCACACGGUGCAGGCAAAUGGACGCUGCUCCAGAUGCACCAAGUUAUGCACCUUUAGGCAGCGUU ((((.((..((((((.((((..(...(((((((.........)))))))...)..)))).))).)))..)).))))((((((((..((.((((........)))).))...)))))))). ( -37.10) >DroSim_CAF1 118219 120 + 1 CUGUAUGUAUGUUGGAGUGCUGCUUCAGCUUCGAAUUGCUGAUGAAGCUCUUGUCACACACUGUGCACGCGAAUGGUCGCUGCUCAAGAUGCACCAAGUUGUGGACCUUAAGACAGCGCU ..........(((((((.(((...(((((........)))))...))).)))(((.(((((((((((.((((....))))..(....).)))))..)).))))))).......))))... ( -34.50) >DroMoj_CAF1 137955 120 + 1 CCGUAUGAAUGUUCGAGUGUUGUUUCAGCUUCGAAUUGCUGAUGAAGCUCUUGUCGCACACGGUACAAGCAAACGGACGCUGCUCCAAGUGCACCAGAUUGUGCACCUUCAGGCAGCGUU ((((.((..((((((.((((..(...(((((((.........)))))))...)..)))).))).)))..)).))))((((((((..((((((((......))))).)))..)))))))). ( -44.50) >consensus CUGUAUGAAUGUUCGAGUGCUGCUUCAGCUUCGAAUUGCUGAUGAAGCUCUUGUCGCACACGGUGCACGCAAAUGGACGCUGCUCAAGAUGCACCAAGUUGUGCACCUUAAGACAGCGCU ((((.((..((((((.((((.((...(((((((.........)))))))...)).)))).))).)))..)).)))).(((((((.(((.(((((......))))).))).)).))))).. (-32.14 = -32.20 + 0.06)

| Location | 24,635,351 – 24,635,471 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.08 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -33.25 |
| Energy contribution | -34.75 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24635351 120 - 27905053 AGCGCUGUCUCAAGGUCCACAACUUGGUGCAUCUUGAGCAGCGACCAUUCGCGUGCACAGUGUGCGACAAGAGCUUCAUCAGCAAUUCGAAGCUUAAGCAGCACUCCAACAUACACACAG .((((((.((((((((.(((......))).))))))))))((((....))))))))...(.((((..(..(((((((...........)))))))..)..)))).).............. ( -39.10) >DroVir_CAF1 131767 120 - 1 AACGCUGCCUAAAGGUGCAUAACUUGGUGCAUCUGGAGCAGCGUCCAUUUGCCUGCACCGUGUGCGACAAGAGCUUCAUUAGCAACUCGAAGCUGAAACAGCACUCGAACAUUCAUACAG .(((((((((..((((((((......)))))))))).)))))))..............((.((((......((((((...........))))))......)))).))............. ( -37.80) >DroSim_CAF1 118219 120 - 1 AGCGCUGUCUUAAGGUCCACAACUUGGUGCAUCUUGAGCAGCGACCAUUCGCGUGCACAGUGUGUGACAAGAGCUUCAUCAGCAAUUCGAAGCUGAAGCAGCACUCCAACAUACAUACAG .((((((.((((((((.(((......))).))))))))))((((....))))))))...((((((....((.(((.(.(((((........))))).).))).))...))))))...... ( -41.10) >DroMoj_CAF1 137955 120 - 1 AACGCUGCCUGAAGGUGCACAAUCUGGUGCACUUGGAGCAGCGUCCGUUUGCUUGUACCGUGUGCGACAAGAGCUUCAUCAGCAAUUCGAAGCUGAAACAACACUCGAACAUUCAUACGG .(((((((((..((((((((......)))))))))).)))))))((((.((..(((.(.((((((.......))....(((((........)))))....))))..).)))..)).)))) ( -42.90) >consensus AACGCUGCCUAAAGGUCCACAACUUGGUGCAUCUGGAGCAGCGACCAUUCGCGUGCACAGUGUGCGACAAGAGCUUCAUCAGCAAUUCGAAGCUGAAACAGCACUCCAACAUACAUACAG ..(((((.((((((((((((......))))))))))))))))).......(..((...((.((((......((((((...........))))))......)))).))..))..)...... (-33.25 = -34.75 + 1.50)


| Location | 24,635,391 – 24,635,511 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -29.24 |
| Energy contribution | -31.43 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24635391 120 + 27905053 GAUGAAGCUCUUGUCGCACACUGUGCACGCGAAUGGUCGCUGCUCAAGAUGCACCAAGUUGUGGACCUUGAGACAGCGCUCCGUUUUGUACAUCUUGCCGCACUUUUCGCAACGAUAGGA ........((((((((((...((((((...((((((.(((((((((((.(.(((......))).).)))))).)))))..))))))))))))...))).((.......))...))))))) ( -42.60) >DroVir_CAF1 131807 120 + 1 AAUGAAGCUCUUGUCGCACACGGUGCAGGCAAAUGGACGCUGCUCCAGAUGCACCAAGUUAUGCACCUUUAGGCAGCGUUCGGUUUUGUACAUUUUGCCGCAUUUCUCGCAGCGAUAGGA ........(((((((((...((((((.(((((((((((((((((..((.((((........)))).))...)))))))))))((.....))..))))))))))....))..))))))))) ( -44.10) >DroSim_CAF1 118259 120 + 1 GAUGAAGCUCUUGUCACACACUGUGCACGCGAAUGGUCGCUGCUCAAGAUGCACCAAGUUGUGGACCUUAAGACAGCGCUCCGUUUUGUACAUCUUGCCGCACUUUUCGCAACGAUAGGA ........(((((((......((((((...((((((.(((((((.(((.(.(((......))).).))).)).)))))..)))))))))))).......((.......))...))))))) ( -33.40) >DroMoj_CAF1 137995 120 + 1 GAUGAAGCUCUUGUCGCACACGGUACAAGCAAACGGACGCUGCUCCAAGUGCACCAGAUUGUGCACCUUCAGGCAGCGUUCGGUUUUGUACAUUUUGCCGCAUUUCUCACAGCGAUAAGA ........((((((((((....(((((((....(((((((((((..((((((((......))))).)))..)))))))))))..)))))))....))).((..........))))))))) ( -46.20) >consensus GAUGAAGCUCUUGUCGCACACGGUGCACGCAAAUGGACGCUGCUCAAGAUGCACCAAGUUGUGCACCUUAAGACAGCGCUCCGUUUUGUACAUCUUGCCGCACUUCUCGCAACGAUAGGA ........(((((((((.....(((((...((((((((((((((.(((.(((((......))))).))).)).)))))))).)))))))))........((.......)).))))))))) (-29.24 = -31.43 + 2.19)



| Location | 24,635,391 – 24,635,511 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -40.05 |
| Consensus MFE | -34.06 |
| Energy contribution | -34.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24635391 120 - 27905053 UCCUAUCGUUGCGAAAAGUGCGGCAAGAUGUACAAAACGGAGCGCUGUCUCAAGGUCCACAACUUGGUGCAUCUUGAGCAGCGACCAUUCGCGUGCACAGUGUGCGACAAGAGCUUCAUC .......((((((.....))))))..............(((((((((.((((((((.(((......))).))))))))))))......(((((..(...)..))))).....)))))... ( -42.00) >DroVir_CAF1 131807 120 - 1 UCCUAUCGCUGCGAGAAAUGCGGCAAAAUGUACAAAACCGAACGCUGCCUAAAGGUGCAUAACUUGGUGCAUCUGGAGCAGCGUCCAUUUGCCUGCACCGUGUGCGACAAGAGCUUCAUU .....((((((((.(...(((((((((..((.....)).(.(((((((((..((((((((......)))))))))).))))))).).)))))).)))))))).))))............. ( -39.30) >DroSim_CAF1 118259 120 - 1 UCCUAUCGUUGCGAAAAGUGCGGCAAGAUGUACAAAACGGAGCGCUGUCUUAAGGUCCACAACUUGGUGCAUCUUGAGCAGCGACCAUUCGCGUGCACAGUGUGUGACAAGAGCUUCAUC .......((((((((..(((((......))))).....((..(((((.((((((((.(((......))).))))))))))))).)).)))))((.(((.....)))))...)))...... ( -38.00) >DroMoj_CAF1 137995 120 - 1 UCUUAUCGCUGUGAGAAAUGCGGCAAAAUGUACAAAACCGAACGCUGCCUGAAGGUGCACAAUCUGGUGCACUUGGAGCAGCGUCCGUUUGCUUGUACCGUGUGCGACAAGAGCUUCAUC ((((.(((((((.......))))).....((((((((.((.(((((((((..((((((((......)))))))))).))))))).)).))..)))))).......)).))))........ ( -40.90) >consensus UCCUAUCGCUGCGAAAAAUGCGGCAAAAUGUACAAAACCGAACGCUGCCUAAAGGUCCACAACUUGGUGCAUCUGGAGCAGCGACCAUUCGCGUGCACAGUGUGCGACAAGAGCUUCAUC .......((((((((..(((((......)))))......(.((((((.((((((((((((......)))))))))))))))))).).))))).(((((...))))).....)))...... (-34.06 = -34.62 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:29:36 2006