
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,630,799 – 24,630,906 |
| Length | 107 |
| Max. P | 0.870339 |

| Location | 24,630,799 – 24,630,906 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.09 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24630799 107 + 27905053 ---AAUCACAAUCACAGGUAACUGCUACAUGUGUAAAUAUUUACAAAACGCGCCAA--GCGUCGACAAACCGACGCCAGG---CAUCUUCACUUUUGCCGCCAACGAGCGGU-----CCU ---............(((..((((((...(((((((....)))))....((.....--((((((......))))))..((---((..........))))))))...))))))-----))) ( -28.00) >DroVir_CAF1 127911 102 + 1 -----UAACAAUCACAGGUAACUGCUACAUGUGUAAAUAUUUACAAAACGCGCCAAAAACGUUGGCAAACAGACGCCAGG---CGCCUACACUUGUACCGUUGCUCA----------UCA -----...........((((((.(.((((.(((((..............(((((........((((........))))))---))).))))).))))).))))))..----------... ( -24.26) >DroPse_CAF1 124617 112 + 1 UA------CAAUCACAGGUAACGGCUACAUGUGUAAAUAUUUACAAAACGCGCCAA--GCGUCGGCAAACUGACGCCAGCCGCCAUCUUCACUUUUGCCGCCAUCUAGCGGGCUAUACAC ..------.......((((..(((((.....(((((....)))))...........--(((((((....))))))).)))))..)))).........((((......))))......... ( -28.40) >DroEre_CAF1 117850 110 + 1 UACAAUCACAAUCACAGGUAACUGCUACAUGUGUAAAUAUUUACAAAACGCGCCAA--GCGUCGACAAACCGACGCCAAG---CAUCUUCACUUUUGCCGCCAACGAGCGGU-----GCU ......((((....(((....))).....))))................(((((..--((((((......))))))....---.............((((....)).)))))-----)). ( -26.40) >DroWil_CAF1 122484 103 + 1 AA------CAAUCACAGGUAACAGCUACAUGUGUAAAUAUUUACAAAACGCGCCAA--GCGUCGGCAAACUGACGCCAGG---CAUCUUCACUUUUACCGUUUUCUCUCAC------UCC ..------........(((((..((......(((((....)))))....))(((..--(((((((....)))))))..))---)..........)))))............------... ( -25.50) >DroPer_CAF1 104363 112 + 1 UA------CAAUCACAGGUAACGGCUACAUGUGUAAAUAUUUACAAAACGCGCCAA--GCGUCGGCAAACUGACGCCAGCCGCCAUCUUCACUUUUGCCGCCAUCUAGCGGGCUAUACAC ..------.......((((..(((((.....(((((....)))))...........--(((((((....))))))).)))))..)))).........((((......))))......... ( -28.40) >consensus UA______CAAUCACAGGUAACUGCUACAUGUGUAAAUAUUUACAAAACGCGCCAA__GCGUCGGCAAACUGACGCCAGG___CAUCUUCACUUUUGCCGCCAUCUAGCGG______CCC ................(((((..((......(((((....)))))....)).......(((((((....)))))))..................)))))..................... (-15.78 = -16.08 + 0.31)

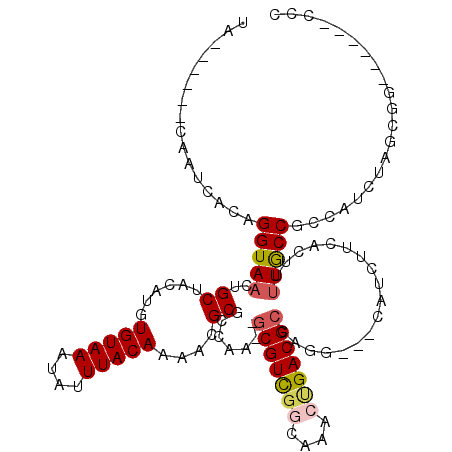

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:29:31 2006