
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,555,023 – 24,555,122 |
| Length | 99 |
| Max. P | 0.750969 |

| Location | 24,555,023 – 24,555,122 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -18.91 |
| Energy contribution | -19.35 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
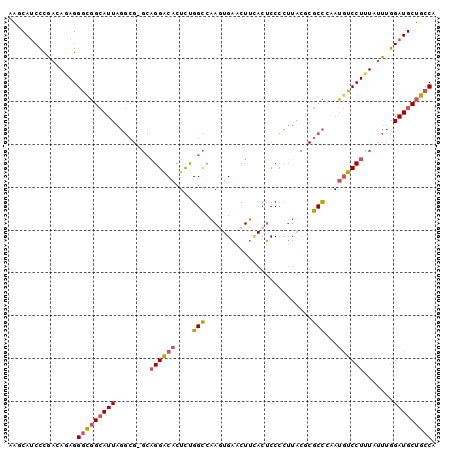
>3R_DroMel_CAF1 24555023 99 + 27905053 AAGCAUCCCGUCACAGGGCGGCAUUAGGCG-GCAGGACACUCUGGCAAAGUGAACUUCACUCCCCUUACGCGCCCAAUGUCCUUUAUUUGGAUGCUGCCA .(((((((.(((....)))((((((.((((-.((((....)))).)..((((.....))))..........))).))))))........))))))).... ( -32.60) >DroSec_CAF1 53234 99 + 1 AAGCAUCCCGACAGAGGGCGGCAUUAGGCG-GCAGGGCACUCUGGCCAAGUGAACUUCACUCCCCUUACGCGCCCAAUGUCCUUUAUUUGGAUGCUGCCA ..((..(((......)))..))....((((-((.((((.(...((...((((.....))))..))....).))))...((((.......)))))))))). ( -37.20) >DroSim_CAF1 55050 100 + 1 AAGCAUCCCAACAGAGGGCGGCAUUAGGCGGGCUGGGCACUCUGGCCAAGUGAACUUCACUCCCCUUACGCGCCCAAUGUCCUUUAUUUGGAUGCUGCCA .(((((((.((.((((((((......(((((((..(.....)..))).((((.....)))).........))))...)))))))).)).))))))).... ( -39.20) >DroEre_CAF1 57673 99 + 1 AAACAUCCUGACAGAGGGUGGCAUUAGGCG-GCAGGACACUCCGGCCAAAUGAACUUAACUCCCUUUACACGCCACAUGUCCUUUAUUUGGAUGCUGCCA ...((((((.....))))))......((((-(((((.....))..(((((((((....((..................))..))))))))).))))))). ( -28.07) >DroYak_CAF1 54487 99 + 1 AAACAUCCUGACAGAGGGCGGCAUUAGGCG-GCAGGACACUCCGGCCAAAUGAACUUUACUCCCCUUACACGCCCCAUGUCCUUUAUUUGGAUGCUGCCA ...(((((.((((..(((((((.....))(-((.((.....)).))).......................)))))..))))........)))))...... ( -29.70) >DroAna_CAF1 54125 79 + 1 ACUCAC-AUGA-AAAGGACUGAA---------AAGGACACUUUGGUCGAGUGUGCUCUACGCC--------GCUCAAAAUCCUUUAUUUU--UGCUGCCG ......-....-((((((((...---------.))(((......)))(((((.((.....)))--------))))....)))))).....--........ ( -18.00) >consensus AAGCAUCCCGACAGAGGGCGGCAUUAGGCG_GCAGGACACUCUGGCCAAGUGAACUUCACUCCCCUUACGCGCCCAAUGUCCUUUAUUUGGAUGCUGCCA ................(((((((((........((((((....(((.........................)))...)))))).......))))))))). (-18.91 = -19.35 + 0.45)


| Location | 24,555,023 – 24,555,122 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -23.61 |
| Energy contribution | -25.58 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.750969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
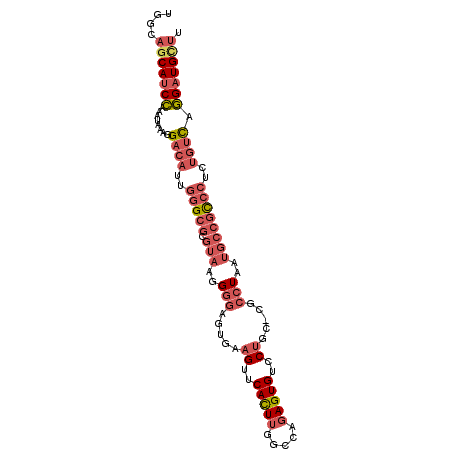
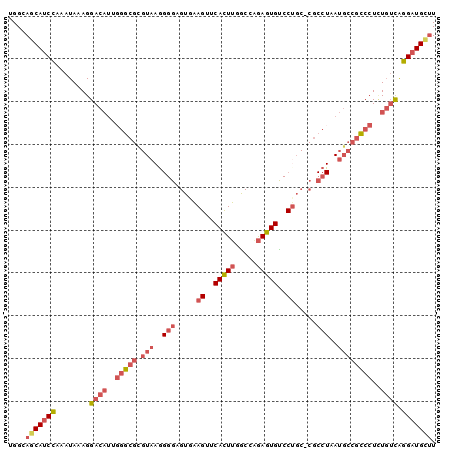
>3R_DroMel_CAF1 24555023 99 - 27905053 UGGCAGCAUCCAAAUAAAGGACAUUGGGCGCGUAAGGGGAGUGAAGUUCACUUUGCCAGAGUGUCCUGC-CGCCUAAUGCCGCCCUGUGACGGGAUGCUU ....(((((((..(((..((.(((((((((.(((.(((.((..(((....)))..).....).))))))-)))))))))))....)))....))))))). ( -37.00) >DroSec_CAF1 53234 99 - 1 UGGCAGCAUCCAAAUAAAGGACAUUGGGCGCGUAAGGGGAGUGAAGUUCACUUGGCCAGAGUGCCCUGC-CGCCUAAUGCCGCCCUCUGUCGGGAUGCUU ....(((((((..(((.(((.(((((((((.(((.((((((((.....))))).((....)).))))))-)))))))))....))).)))..))))))). ( -40.90) >DroSim_CAF1 55050 100 - 1 UGGCAGCAUCCAAAUAAAGGACAUUGGGCGCGUAAGGGGAGUGAAGUUCACUUGGCCAGAGUGCCCAGCCCGCCUAAUGCCGCCCUCUGUUGGGAUGCUU ....(((((((.((((.(((.((((((((.(....)(((((((.....)))).(((......)))...)))))))))))....))).)))).))))))). ( -39.10) >DroEre_CAF1 57673 99 - 1 UGGCAGCAUCCAAAUAAAGGACAUGUGGCGUGUAAAGGGAGUUAAGUUCAUUUGGCCGGAGUGUCCUGC-CGCCUAAUGCCACCCUCUGUCAGGAUGUUU ....(((((((..(((.(((....(((((((....((((((.....)))....(((.((.....)).))-).))).)))))))))).)))..))))))). ( -32.60) >DroYak_CAF1 54487 99 - 1 UGGCAGCAUCCAAAUAAAGGACAUGGGGCGUGUAAGGGGAGUAAAGUUCAUUUGGCCGGAGUGUCCUGC-CGCCUAAUGCCGCCCUCUGUCAGGAUGUUU ....(((((((........((((.((((((.(((..(((...(((.....)))(((.((.....)).))-).)))..))))))))).)))).))))))). ( -36.00) >DroAna_CAF1 54125 79 - 1 CGGCAGCA--AAAAUAAAGGAUUUUGAGC--------GGCGUAGAGCACACUCGACCAAAGUGUCCUU---------UUCAGUCCUUU-UCAU-GUGAGU ......((--.....((((((((..(((.--------(((.....)).).)))(((......)))...---------...))))))))-....-.))... ( -15.40) >consensus UGGCAGCAUCCAAAUAAAGGACAUUGGGCGCGUAAGGGGAGUGAAGUUCACUUGGCCAGAGUGUCCUGC_CGCCUAAUGCCGCCCUCUGUCAGGAUGCUU ....(((((((........((((..(((((.(((..(((.....((..(((((.....)))))..)).....)))..))))))))..)))).))))))). (-23.61 = -25.58 + 1.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:29:07 2006