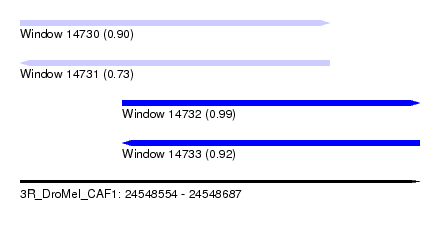
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,548,554 – 24,548,687 |
| Length | 133 |
| Max. P | 0.988579 |

| Location | 24,548,554 – 24,548,657 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.63 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.94 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24548554 103 + 27905053 ACUUCCCC-CCAGAAGUAUGCAU--ACUUGCACUCCG------CUCGCACUUUGUCG----CCCCA-ACCGGGCGGCAAAGCCAAUCCAGUUCUGGCUUU-G-CCCAACCGACAAGCGC (((((...-...))))).((((.--...)))).....------..(((...((((((----(((..-...))))((((((((((.........)))))))-)-)).....)))))))). ( -38.40) >DroSec_CAF1 46792 103 + 1 ACUUCCCC-CCAGAAGUAUGCAU--ACUUGCACUCCG------CUCACACUUUGUCG----CCCUA-ACCGGGCUGCAAAGCCAAUCCAGUUCUGGCUUU-G-CCCAACCGACAAGCGC (((((...-...))))).((((.--...))))...((------((.......(((((----(((..-...)))).(((((((((.........)))))))-)-)......)))))))). ( -34.31) >DroSim_CAF1 48603 103 + 1 ACUUCCCC-CCAGAAGUAUGCAU--ACUUGCACUCCG------CUCACACUUUGUCG----CCCCA-ACCGGGCGGCAAAGCCAAUCCAGUUCUGGCUUU-G-GCCAACCGACAAGCGC (((((...-...))))).((((.--...))))...((------((.....(((((((----(((..-...))))))))))(((((.(((....)))..))-)-)).........)))). ( -36.30) >DroEre_CAF1 51131 103 + 1 ACUUCCUC-CCAUAAGUAUGCAU--ACUUGCACUCCG------CUCGCACUUUGUCG----CCCCA-GCCGGGCGGCAAAGCCAAUCCAGUUCUGGCUUU-G-CCCGACCGACAAGCGA ........-.........((((.--...)))).....------.((((...((((((----(((..-...))))((((((((((.........)))))))-)-)).....))))))))) ( -36.50) >DroYak_CAF1 47896 105 + 1 ACUUCCCCACCAGAAGUAUGCAU--ACUUGCACUCCG------CUCGCACUUUGUCG----CCCCAAACCGGGCGGCAAAGCCAAUCCAGUUCUGGCUUU-G-CCCAACCGACAAGCGC (((((.......))))).((((.--...)))).....------..(((...((((((----(((......))))((((((((((.........)))))))-)-)).....)))))))). ( -38.10) >DroAna_CAF1 46967 114 + 1 AGA---CC-CUUAAAGUAUGCAAGGACUCCUACUCCCCGACUCCUUCCAUUUUACCGGAUUCCCGG-AUCAAGUCGGCAAGCCAAUCCAUUUUUGGCUGUUGUUCUGGCCGACGAGGGC ...---((-(((.....(((.(((((.((.........)).))))).)))....((((....))))-.....((((((.((((((.......)))))).........))))))))))). ( -37.30) >consensus ACUUCCCC_CCAGAAGUAUGCAU__ACUUGCACUCCG______CUCGCACUUUGUCG____CCCCA_ACCGGGCGGCAAAGCCAAUCCAGUUCUGGCUUU_G_CCCAACCGACAAGCGC .........((((((.(.((((......)))).................((((((((....(((......))))))))))).......).))))))....................... (-14.88 = -15.63 + 0.75)



| Location | 24,548,554 – 24,548,657 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -22.21 |
| Energy contribution | -23.77 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24548554 103 - 27905053 GCGCUUGUCGGUUGGG-C-AAAGCCAGAACUGGAUUGGCUUUGCCGCCCGGU-UGGGG----CGACAAAGUGCGAG------CGGAGUGCAAGU--AUGCAUACUUCUGG-GGGGAAGU .(((((((...(((((-(-((((((((.......)))))))))))((((...-..)))----)..)))...)))))------))..(((((...--.)))))((((((..-..)))))) ( -46.50) >DroSec_CAF1 46792 103 - 1 GCGCUUGUCGGUUGGG-C-AAAGCCAGAACUGGAUUGGCUUUGCAGCCCGGU-UAGGG----CGACAAAGUGUGAG------CGGAGUGCAAGU--AUGCAUACUUCUGG-GGGGAAGU (((((((((......(-(-((((((((.......)))))))))).((((...-..)))----)))).))))))...------....(((((...--.)))))((((((..-..)))))) ( -41.10) >DroSim_CAF1 48603 103 - 1 GCGCUUGUCGGUUGGC-C-AAAGCCAGAACUGGAUUGGCUUUGCCGCCCGGU-UGGGG----CGACAAAGUGUGAG------CGGAGUGCAAGU--AUGCAUACUUCUGG-GGGGAAGU ((((((((((((.(((-(-((..(((....))).))))))..)))((((...-..)))----)))).))))))...------....(((((...--.)))))((((((..-..)))))) ( -41.70) >DroEre_CAF1 51131 103 - 1 UCGCUUGUCGGUCGGG-C-AAAGCCAGAACUGGAUUGGCUUUGCCGCCCGGC-UGGGG----CGACAAAGUGCGAG------CGGAGUGCAAGU--AUGCAUACUUAUGG-GAGGAAGU (((((((((.(((.((-(-((((((((.......)))))))))))((((...-..)))----))))...).)))))------))).(((((...--.))))).(((....-)))..... ( -44.20) >DroYak_CAF1 47896 105 - 1 GCGCUUGUCGGUUGGG-C-AAAGCCAGAACUGGAUUGGCUUUGCCGCCCGGUUUGGGG----CGACAAAGUGCGAG------CGGAGUGCAAGU--AUGCAUACUUCUGGUGGGGAAGU .(((((((...(((((-(-((((((((.......)))))))))))((((......)))----)..)))...)))))------))..(((((...--.)))))((((((.....)))))) ( -46.70) >DroAna_CAF1 46967 114 - 1 GCCCUCGUCGGCCAGAACAACAGCCAAAAAUGGAUUGGCUUGCCGACUUGAU-CCGGGAAUCCGGUAAAAUGGAAGGAGUCGGGGAGUAGGAGUCCUUGCAUACUUUAAG-GG---UCU (((((.((((((.((.....((((((....))).))).)).)))))).....-((((....))))....(((.(((((.((.........)).))))).)))......))-))---).. ( -38.30) >consensus GCGCUUGUCGGUUGGG_C_AAAGCCAGAACUGGAUUGGCUUUGCCGCCCGGU_UGGGG____CGACAAAGUGCGAG______CGGAGUGCAAGU__AUGCAUACUUCUGG_GGGGAAGU ((((((((((.........((((((((.......))))))))....(((......)))....)))).)))))).............(((((......)))))((((((.....)))))) (-22.21 = -23.77 + 1.56)



| Location | 24,548,588 – 24,548,687 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -18.93 |
| Energy contribution | -20.82 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24548588 99 + 27905053 CUCGCACUUUGUCGCCCCA-ACCGGGCGGCAAAGCCAAUCCAGUUCUGGCUUUGCCCAACCGACAAGCGCACAAACAUCAUAACCACAUCAGCA--------CGUGAA ...((.(((.(((((((..-...))))((((((((((.........)))))))))).....)))))).))..............(((.......--------.))).. ( -34.70) >DroVir_CAF1 41501 82 + 1 CUUUGCCCUUAAAGCGCCA-UACGC---------GCAAUCCAUUUUUCGUA--G----CUUGGCAAAAGCGCAAACAUCAAAACCACAUCAGCG--------GC--AU ...((((((....((((..-...))---------))............((.--(----(((.....))))))..................)).)--------))--). ( -16.70) >DroSec_CAF1 46826 99 + 1 CUCACACUUUGUCGCCCUA-ACCGGGCUGCAAAGCCAAUCCAGUUCUGGCUUUGCCCAACCGACAAGCGCACAAACAUCAUAACCACAUCAGCA--------CGUGAA .((((...(((((((((..-...)))).(((((((((.........)))))))))......)))))((.......................)).--------.)))). ( -30.50) >DroSim_CAF1 48637 99 + 1 CUCACACUUUGUCGCCCCA-ACCGGGCGGCAAAGCCAAUCCAGUUCUGGCUUUGGCCAACCGACAAGCGCACAAACAUCAUAACCACAUCAGCA--------CGUGAA .((((...(((((((((..-...))))((((((((((.........))))))).)))....)))))((.......................)).--------.)))). ( -31.20) >DroEre_CAF1 51165 107 + 1 CUCGCACUUUGUCGCCCCA-GCCGGGCGGCAAAGCCAAUCCAGUUCUGGCUUUGCCCGACCGACAAGCGAACAAACAUCAUAACCACAUCAGCAGAGGAGCAGGGGAG .((((...(((((((((..-...))))((((((((((.........)))))))))).....))))))))).............((.(....((......))..))).. ( -35.90) >DroYak_CAF1 47931 100 + 1 CUCGCACUUUGUCGCCCCAAACCGGGCGGCAAAGCCAAUCCAGUUCUGGCUUUGCCCAACCGACAAGCGCACAAACAUCAUAACCACAUCAGCA--------GGGGAA ...((.(((.(((((((......))))((((((((((.........)))))))))).....)))))).)).......((....((.........--------)).)). ( -33.60) >consensus CUCGCACUUUGUCGCCCCA_ACCGGGCGGCAAAGCCAAUCCAGUUCUGGCUUUGCCCAACCGACAAGCGCACAAACAUCAUAACCACAUCAGCA________CGUGAA ........(((((((((......))))((((((((((.........)))))))))).....))))).......................................... (-18.93 = -20.82 + 1.89)



| Location | 24,548,588 – 24,548,687 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.78 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24548588 99 - 27905053 UUCACG--------UGCUGAUGUGGUUAUGAUGUUUGUGCGCUUGUCGGUUGGGCAAAGCCAGAACUGGAUUGGCUUUGCCGCCCGGU-UGGGGCGACAAAGUGCGAG .(((((--------(....))))))............((((((((((.....(((((((((((.......)))))))))))((((...-..))))))).))))))).. ( -42.20) >DroVir_CAF1 41501 82 - 1 AU--GC--------CGCUGAUGUGGUUUUGAUGUUUGCGCUUUUGCCAAG----C--UACGAAAAAUGGAUUGC---------GCGUA-UGGCGCUUUAAGGGCAAAG .(--((--------(.(((((.(..(((((..(((((.((....))))))----)--..)))))...).)))((---------((...-..))))....))))))... ( -22.40) >DroSec_CAF1 46826 99 - 1 UUCACG--------UGCUGAUGUGGUUAUGAUGUUUGUGCGCUUGUCGGUUGGGCAAAGCCAGAACUGGAUUGGCUUUGCAGCCCGGU-UAGGGCGACAAAGUGUGAG .(((((--------(((.(((((.......)))))...))))(((((......((((((((((.......)))))))))).((((...-..)))))))))...)))). ( -38.00) >DroSim_CAF1 48637 99 - 1 UUCACG--------UGCUGAUGUGGUUAUGAUGUUUGUGCGCUUGUCGGUUGGCCAAAGCCAGAACUGGAUUGGCUUUGCCGCCCGGU-UGGGGCGACAAAGUGUGAG .(((((--------(((.(((((.......)))))...))))((((((((.((((((..(((....))).))))))..)))((((...-..)))))))))...)))). ( -38.60) >DroEre_CAF1 51165 107 - 1 CUCCCCUGCUCCUCUGCUGAUGUGGUUAUGAUGUUUGUUCGCUUGUCGGUCGGGCAAAGCCAGAACUGGAUUGGCUUUGCCGCCCGGC-UGGGGCGACAAAGUGCGAG ......(((.(..(..(....)..)........(((((.((((.(((((.(((.(((((((((.......)))))))))))).)))))-...)))))))))).))).. ( -37.70) >DroYak_CAF1 47931 100 - 1 UUCCCC--------UGCUGAUGUGGUUAUGAUGUUUGUGCGCUUGUCGGUUGGGCAAAGCCAGAACUGGAUUGGCUUUGCCGCCCGGUUUGGGGCGACAAAGUGCGAG .....(--------..(....)..)............((((((((((.....(((((((((((.......)))))))))))((((......))))))).))))))).. ( -40.10) >consensus UUCACC________UGCUGAUGUGGUUAUGAUGUUUGUGCGCUUGUCGGUUGGGCAAAGCCAGAACUGGAUUGGCUUUGCCGCCCGGU_UGGGGCGACAAAGUGCGAG ...............((.(((((.......))))).))(((((((((.....(((((((((((.......)))))))))))((((......))))))).))))))... (-25.64 = -26.78 + 1.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:29:02 2006