| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
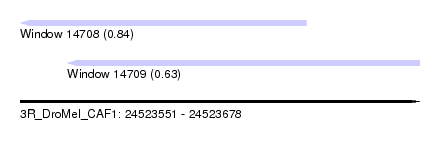
| Location | 24,523,551 – 24,523,678 |
| Length | 127 |
| Max. P | 0.836812 |

| Location | 24,523,551 – 24,523,642 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.95 |
| Mean single sequence MFE | -19.39 |
| Consensus MFE | -14.76 |
| Energy contribution | -15.80 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
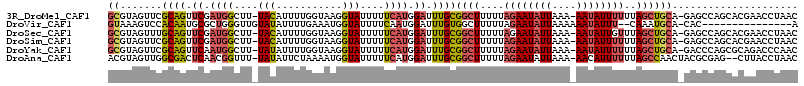
>3R_DroMel_CAF1 24523551 91 - 27905053 AGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAAAAUAUUUUUUAGCUGCA-GAGCCAGCACGAACCUAACAAUAUAUGUAUAUAU ((((........(((.(((((((((....(((((((.....)))))))..)))))))-)).)))......)))).................. ( -22.34) >DroSec_CAF1 21956 91 - 1 AGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAAAAUAUUGUUUAGCUGCA-GAGCCAGCACGAACCUAACAAUAUAUGUAUAUAU ((((........(((.(((((((((....(.(((((.....))))).)..)))))))-)).)))......)))).................. ( -20.84) >DroSim_CAF1 23957 91 - 1 AGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAAAAUAUUUUUUAGCUGCA-GAGCCAGCACGAACCUAACAAUAUAUGUAUAUAU ((((........(((.(((((((((....(((((((.....)))))))..)))))))-)).)))......)))).................. ( -22.34) >DroYak_CAF1 22345 91 - 1 AGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAAAAUAUUUUUUAGCUGCA-GACCCAGCGCAGACCCAACAAUAUUUAUAUAUAU .(((........(((.(((((((((....(((((((.....)))))))..)))))))-)).)))......)))................... ( -22.14) >DroAna_CAF1 22762 86 - 1 UGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAAAACAUUUUUUAGCCAACUACGCGAG--CUUACCUAACAAUAUAUGUA----U .((((...(((.((.......((((....(((((.........)))))..)))).....)).)))--..))))..............----. ( -9.30) >consensus AGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAAAAUAUUUUUUAGCUGCA_GAGCCAGCACGAACCUAACAAUAUAUGUAUAUAU .(((........(((...(((((((....(((((((.....)))))))..)))))))....)))......)))................... (-14.76 = -15.80 + 1.04)



| Location | 24,523,566 – 24,523,678 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -30.51 |
| Consensus MFE | -10.69 |
| Energy contribution | -11.25 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24523566 112 - 27905053 GCGUAGUUCGCAGUUCGAUGGCUU-UACAUUUUGGUAAGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAA-AAUAUUUUUUAGCUGCA-GAGCCAGCACGAACCUAAC (((.....))).(((((.((((((-(((......)))))))........(((.(((((((((....(((((((....-.)))))))..)))))))-)).))).)))))))..... ( -35.20) >DroVir_CAF1 10841 97 - 1 GUAAAGUCCACAAUGCGCUGGGUUGUAUAUUUUGAAAUGGUAUUUUCAAUGGAUUUGUGGCUUUUUAGAAUAUUAAAAAAUAUUU--CAAAUGCA-CAC---------------A ...(((((((((((.(...).))))).....((((((......)))))).))))))(((((..(((..((((((....)))))).--.))).)).-)))---------------. ( -20.70) >DroSec_CAF1 21971 112 - 1 GCGUAGUUUGCAGUUCGAUGGCUU-UACAUUUUGGUAAGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAA-AAUAUUGUUUAGCUGCA-GAGCCAGCACGAACCUAAC (((.....))).(((((.((((((-(((......)))))))........(((.(((((((((....(.(((((....-.))))).)..)))))))-)).))).)))))))..... ( -32.30) >DroSim_CAF1 23972 112 - 1 GCGUAGUUCGCAGUUCGAUGGCUU-UACAUUUUGGUAAGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAA-AAUAUUUUUUAGCUGCA-GAGCCAGCACGAACCUAAC (((.....))).(((((.((((((-(((......)))))))........(((.(((((((((....(((((((....-.)))))))..)))))))-)).))).)))))))..... ( -35.20) >DroYak_CAF1 22360 112 - 1 GCGUAGUUCGCAGUUCAAUGGCUU-UAUAUUUUGGUAAGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAA-AAUAUUUUUUAGCUGCA-GACCCAGCGCAGACCCAAC ((((................((((-(((......)))))))........(((.(((((((((....(((((((....-.)))))))..)))))))-)).)))))))......... ( -29.80) >DroAna_CAF1 22773 111 - 1 ACGUAGUUGGCGACUCAACGGUUU-UAUAUUCUAAAAUGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAA-AACAUUUUUUAGCCAACUACGCGAG--CUUACCUAAC .((((((((((.........((((-(((((((((((((.(((..((.....))..))).)..)))))))))))..))-)))........))))))))))....--.......... ( -29.83) >consensus GCGUAGUUCGCAGUUCGAUGGCUU_UACAUUUUGGUAAGGUAUUUUUCAUGGAUUUGCGGCUUUUUAGAAUAUUAAA_AAUAUUUUUUAGCUGCA_GAGCCAGCACGAACCUAAC ((.......((((.((.((((....(((...........)))....)))).)).))))((((....((((((((....))))))))..))))))..................... (-10.69 = -11.25 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:28:42 2006