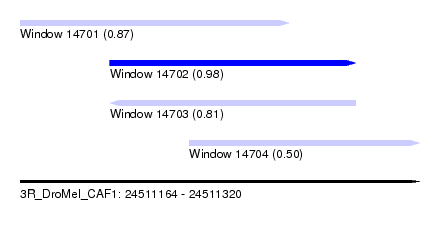
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,511,164 – 24,511,320 |
| Length | 156 |
| Max. P | 0.983302 |

| Location | 24,511,164 – 24,511,269 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -21.43 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24511164 105 + 27905053 CUUUCCGCUGCACCGGAAA--CUCACGGAGCU--UCACCAUUGCAUGCUAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAU ......(((((.(((....--....))).)).--......(((((((((.((((..........))))..........((....)).....)))))))))....))).. ( -28.50) >DroPse_CAF1 11397 99 + 1 CUUUCCGUUGCAUCGGAAAGGCCCCCGGAAAGCCAUUGCAUUGCAUGCCA-CCACGACUGACCUUGUC---------ACGGCAACGCACUCGGCAUGCAAACUACAGAU (((((((......)))))))((....((....))...)).(((((((((.-....(((.......)))---------.((....)).....)))))))))......... ( -32.50) >DroSim_CAF1 9857 105 + 1 CUUUCCGCUGCACCGGAAA--CUCACGGAGCU--UCACCAUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAU ......(((((.(((....--....))).)).--......(((((((((.((((..........))))..........((....)).....)))))))))....))).. ( -31.20) >DroEre_CAF1 9766 105 + 1 CUUUCCGCUGCACCGGAAA--CUCACGGAGCA--UCACCAUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAU ......(((((.(((....--....))).)).--......(((((((((.((((..........))))..........((....)).....)))))))))....))).. ( -31.70) >DroYak_CAF1 9892 105 + 1 CUUUCCGCUGCACCGGAAA--CUCACGGAGCU--ACACCAUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAU ......(((((.(((....--....))).)).--......(((((((((.((((..........))))..........((....)).....)))))))))....))).. ( -31.20) >DroAna_CAF1 10375 88 + 1 UUUUCCGCUGCUCGGGAAA--GCACCGGAAAG-----ACAUUGCAUGCCAGCUUCUAUUGACCUUGGC--------------AACGCACUCGGCAUGCAAACUAAGCAU (((((((.((((......)--))).)))))))-----...(((((((((....((....))....(..--------------..)......)))))))))......... ( -28.50) >consensus CUUUCCGCUGCACCGGAAA__CUCACGGAGCU__UCACCAUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAU .((((((......)))))).....................(((((((((.((((..........))))..........((....)).....)))))))))......... (-21.43 = -22.15 + 0.72)

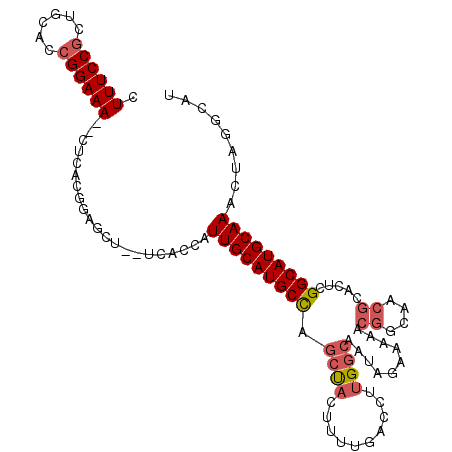
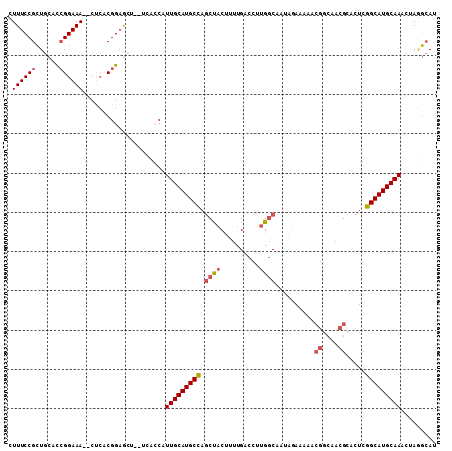
| Location | 24,511,199 – 24,511,295 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -18.76 |
| Energy contribution | -18.79 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24511199 96 + 27905053 AUUGCAUGCUAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCGAAUAGCAAAUG--------------AUUGCUAAGA .(((((((((.((((..........))))..........((....)).....))))))))).((.(((((((((.....))...))--------------).)))).)). ( -26.10) >DroPse_CAF1 11436 100 + 1 AUUGCAUGCCA-CCACGACUGACCUUGUC---------ACGGCAACGCACUCGGCAUGCAAACUACAGAUCAGCAAAACCUCAACGAUCCGUAGUAGCGUUUUGCUAAGA .(((((((((.-....(((.......)))---------.((....)).....)))))))))..........(((((((((((.(((...))).).)).)))))))).... ( -32.70) >DroSec_CAF1 9848 96 + 1 AUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCGAAUAGCAGAUG--------------AUUGCUAAGA .(((((((((.((((..........))))..........((....)).....))))))))).((.(((((((((.....))...))--------------).)))).)). ( -28.80) >DroEre_CAF1 9801 96 + 1 AUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCAAAUAGCAAAUG--------------AUUGCUAAGA .(((((((((.((((..........))))..........((....)).....))))))))).((.(((((((((.....))...))--------------).)))).)). ( -28.10) >DroYak_CAF1 9927 96 + 1 AUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCGAAUAGCAAAUG--------------AUUGCUAAGA .(((((((((.((((..........))))..........((....)).....))))))))).((.(((((((((.....))...))--------------).)))).)). ( -28.80) >DroAna_CAF1 10407 75 + 1 AUUGCAUGCCAGCUUCUAUUGACCUUGGC--------------AACGCACUCGGCAUGCAAACUAAGCAUCAACAAUCAG---------------------UUGUUAAGA .(((((((((....((....))....(..--------------..)......))))))))).((.((((.(........)---------------------.)))).)). ( -18.50) >consensus AUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCAAAUAGCAAAUG______________AUUGCUAAGA .(((((((((.((((..........))))..........((....)).....)))))))))..........(((((.........................))))).... (-18.76 = -18.79 + 0.03)

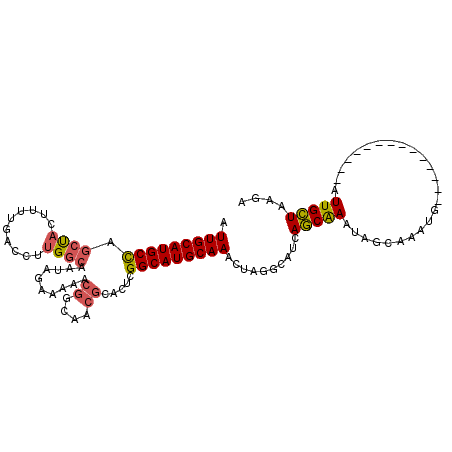
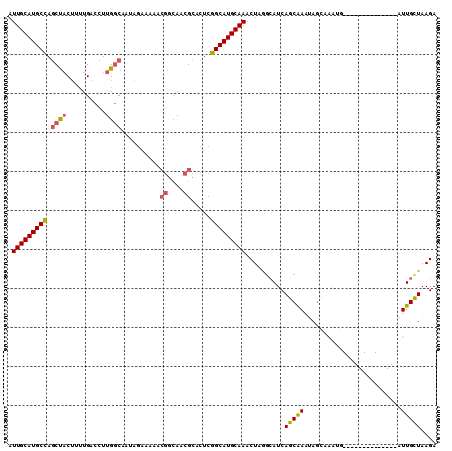
| Location | 24,511,199 – 24,511,295 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -17.59 |
| Energy contribution | -18.01 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24511199 96 - 27905053 UCUUAGCAAU--------------CAUUUGCUAUUCGCUGAUGCCUAGUUUGCAUGCCGAGUGCGUUGCCGUUUUUCUAUUGCCAAGGUCAAAAGUAGCUAGCAUGCAAU ...((((((.--------------...))))))...((....)).....((((((((..(((...(((((................)).))).....))).)))))))). ( -22.69) >DroPse_CAF1 11436 100 - 1 UCUUAGCAAAACGCUACUACGGAUCGUUGAGGUUUUGCUGAUCUGUAGUUUGCAUGCCGAGUGCGUUGCCGU---------GACAAGGUCAGUCGUGG-UGGCAUGCAAU ...((((.....))))(((((((((((.........)).))))))))).((((((((((..(((((((((..---------.....)).))).)))).-)))))))))). ( -37.50) >DroSec_CAF1 9848 96 - 1 UCUUAGCAAU--------------CAUCUGCUAUUCGCUGAUGCCUAGUUUGCAUGCCGAGUGCGUUGCCGUUUUUCUAUUGCCAAGGUCAAAAGUAGCUGGCAUGCAAU ..((((....--------------((((.((.....)).)))).)))).((((((((((..(((.(((((................)).)))..)))..)))))))))). ( -25.59) >DroEre_CAF1 9801 96 - 1 UCUUAGCAAU--------------CAUUUGCUAUUUGCUGAUGCCUAGUUUGCAUGCCGAGUGCGUUGCCGUUUUUCUAUUGCCAAGGUCAAAAGUAGCUGGCAUGCAAU ...((((((.--------------...))))))...((....)).....((((((((((..(((.(((((................)).)))..)))..)))))))))). ( -26.49) >DroYak_CAF1 9927 96 - 1 UCUUAGCAAU--------------CAUUUGCUAUUCGCUGAUGCCUAGUUUGCAUGCCGAGUGCGUUGCCGUUUUUCUAUUGCCAAGGUCAAAAGUAGCUGGCAUGCAAU ...((((((.--------------...))))))...((....)).....((((((((((..(((.(((((................)).)))..)))..)))))))))). ( -26.79) >DroAna_CAF1 10407 75 - 1 UCUUAACAA---------------------CUGAUUGUUGAUGCUUAGUUUGCAUGCCGAGUGCGUU--------------GCCAAGGUCAAUAGAAGCUGGCAUGCAAU ......(((---------------------(.....)))).........(((((((((.(((..(((--------------((....).))))....)))))))))))). ( -22.80) >consensus UCUUAGCAAU______________CAUUUGCUAUUCGCUGAUGCCUAGUUUGCAUGCCGAGUGCGUUGCCGUUUUUCUAUUGCCAAGGUCAAAAGUAGCUGGCAUGCAAU ..(((((.............................)))))........((((((((((..(((...(((................))).....)))..)))))))))). (-17.59 = -18.01 + 0.42)

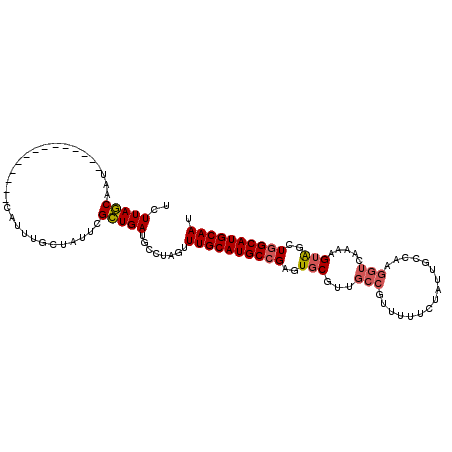
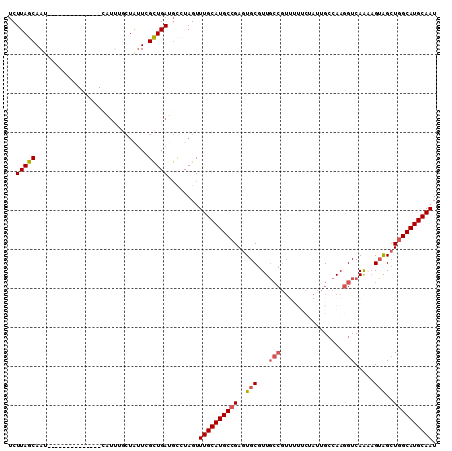
| Location | 24,511,230 – 24,511,320 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.22 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.97 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24511230 90 + 27905053 UAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCGAAUAGCAAAUGAUUGCUAAGAACUCGCCUGUUGUUUUCAAAGCUGA ........((....))...(((((.((.(((((((((.........((((((....)))))).(....))))))..)))).))..))))) ( -20.70) >DroSec_CAF1 9879 90 + 1 UAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCGAAUAGCAGAUGAUUGCUAAGAACUCGCCUGUUGUUUUCAAAGCUGA ........((....))...(((((((((.......))))..(.(((((((((.(((.(.......).))).))))))))).)...))))) ( -20.00) >DroSim_CAF1 9923 90 + 1 UAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCGAAUAGCAAAUGAUUGCUAAGAACUCGCCUGUUGUUUUCAAAGCUGA ........((....))...(((((.((.(((((((((.........((((((....)))))).(....))))))..)))).))..))))) ( -20.70) >DroEre_CAF1 9832 90 + 1 UAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCAAAUAGCAAAUGAUUGCUAAGAACUCGCCUGUUGUUUUCAAAGCUGA ........((....))...(((((.((.(((((((((.........((((((....)))))).(....))))))..)))).))..))))) ( -20.70) >DroYak_CAF1 9958 90 + 1 UAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCGAAUAGCAAAUGAUUGCUAAGAACUUGCCUGUUGUUUUCAAAGCUGA ........((....))...(((((.((.((((((((((....(...((((((....)))))).)....))))))..)))).))..))))) ( -22.90) >consensus UAGAAAAACGGCAACGCACUCGGCAUGCAAACUAGGCAUCAGCGAAUAGCAAAUGAUUGCUAAGAACUCGCCUGUUGUUUUCAAAGCUGA ........((....))...(((((.((.(((((((((.....(...((((((....)))))).).....)))))..)))).))..))))) (-20.38 = -20.22 + -0.16)


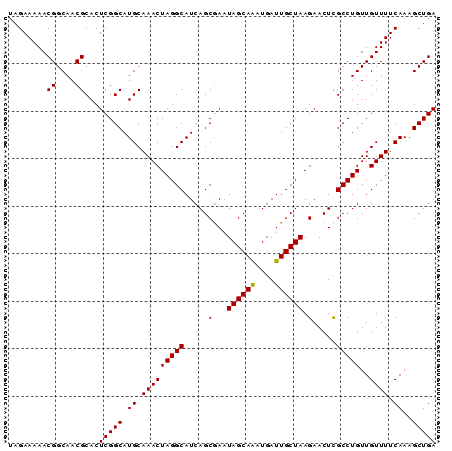
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:28:37 2006