
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
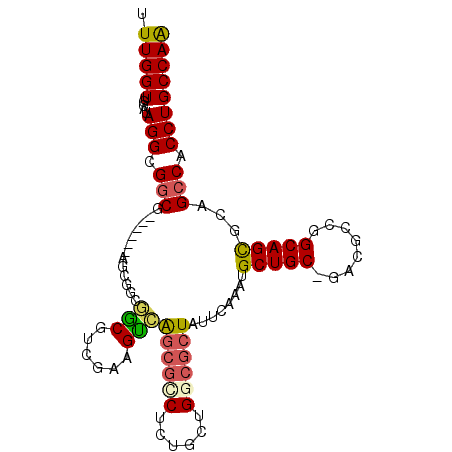
| Location | 3,108,199 – 3,108,294 |
| Length | 95 |
| Max. P | 0.860929 |

| Location | 3,108,199 – 3,108,294 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -46.35 |
| Consensus MFE | -28.03 |
| Energy contribution | -27.95 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
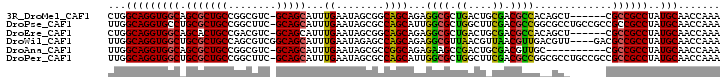
>3R_DroMel_CAF1 3108199 95 + 27905053 UUUGGUUGCAUAGGCGGCG------AGCUGUGGCGUCGCAGUCAGCGCCUCUGCUGCCGCUAUUCAAAUGCUGC-GACGCCGGCAGCGCUGCCACCUGCCAG ..((((.....(((.((((------.(((((((((((((((((((((....))))).............)))))-)))))).)))))..)))).))))))). ( -47.71) >DroPse_CAF1 31371 101 + 1 UUUGGUUGCAUAGGCGGCGGCGGCAGGCGCCGGCGUCGAAGCCAGCGCCAAUGCUGGCGCUAUUCAAAUGCUGC-GAAGCCGGCAGCGCAGGCACCUGCCAA ....(((((....)))))...((((((.((((((......)))(((((((....)))))))........(((((-.......)))))...))).)))))).. ( -51.50) >DroEre_CAF1 34618 95 + 1 UUUGGUUGCAUAGGCGGCG------AGCUGUGGCGUCGCAGUCAGCGCCUCUGCUGCCGCUAUUCAAAUGCUGC-GACGUCGGCAGUGCUGCCACCUGCCAG ..((((.....(((.((((------.(((((((((((((((((((((....))))).............)))))-)))))).)))))..)))).))))))). ( -43.41) >DroWil_CAF1 48731 98 + 1 UUUGGUUGCAUAGGCGGCGUC----AACGUCAACGUUAACGUUAACGCCUCUGCUGGCUCUAUUCAAAUGCUGCCGACGCUGGCAGCGCAGCCACCUGCCAA ..((((((((((((((((((.----(((((........))))).))))).(....))).))))......(((((((....))))))))))))))........ ( -39.50) >DroAna_CAF1 34388 91 + 1 UUUGGUUGCAUAGGCGGCG----------GCAACGUCGCAGUCGGCUUCUCUGCCGGCGCUAUUCAAAUGCUGC-GACGCCGGCAGCGCUGCCACCUGCCAA .(((((.....(((.((((----------((...((((((((((((......))))))((.........)))))-)))((.....)))))))).)))))))) ( -43.50) >DroPer_CAF1 31340 101 + 1 UUUGGUUGCAUAGGCGGCGGCGGCAGGCGCCGGCGUCGAAGCCAGCGCCAAUGCUGGCGCUAUUCAAAUGCUGC-GAAGCCGGCAGCGCAGCCACCUGCCAA ..(((((((...((((.(((((...(((((.(((......))).)))))..))))).))))........(((((-.......))))))))))))........ ( -52.50) >consensus UUUGGUUGCAUAGGCGGCG______AGCGGCGGCGUCGAAGUCAGCGCCUCUGCUGGCGCUAUUCAAAUGCUGC_GACGCCGGCAGCGCAGCCACCUGCCAA .(((((.....(((.(((.............(((......)))((((((......))))))........(((((........)))))...))).)))))))) (-28.03 = -27.95 + -0.08)

| Location | 3,108,199 – 3,108,294 |
|---|---|
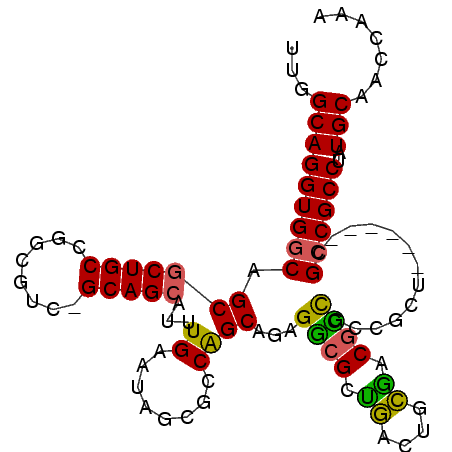
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -46.17 |
| Consensus MFE | -27.66 |
| Energy contribution | -26.88 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3108199 95 - 27905053 CUGGCAGGUGGCAGCGCUGCCGGCGUC-GCAGCAUUUGAAUAGCGGCAGCAGAGGCGCUGACUGCGACGCCACAGCU------CGCCGCCUAUGCAACCAAA ...(((((((((.(.((((..((((((-(((((....)..(((((.(......).))))).)))))))))).)))).------)))))))..)))....... ( -47.50) >DroPse_CAF1 31371 101 - 1 UUGGCAGGUGCCUGCGCUGCCGGCUUC-GCAGCAUUUGAAUAGCGCCAGCAUUGGCGCUGGCUUCGACGCCGGCGCCUGCCGCCGCCGCCUAUGCAACCAAA ..((((((((((.(((((((.......-)))))..((((((((((((......)))))))..))))).)).))))))))))......((....))....... ( -49.90) >DroEre_CAF1 34618 95 - 1 CUGGCAGGUGGCAGCACUGCCGACGUC-GCAGCAUUUGAAUAGCGGCAGCAGAGGCGCUGACUGCGACGCCACAGCU------CGCCGCCUAUGCAACCAAA ...((((((((((((..((..(.((((-(((((....)..(((((.(......).))))).)))))))))..)))))------.))))))..)))....... ( -37.20) >DroWil_CAF1 48731 98 - 1 UUGGCAGGUGGCUGCGCUGCCAGCGUCGGCAGCAUUUGAAUAGAGCCAGCAGAGGCGUUAACGUUAACGUUGACGUU----GACGCCGCCUAUGCAACCAAA ..(((.(.(((((..((((((......)))))).((.....))))))).)...(((((((((((((....)))))))----)))))))))............ ( -44.10) >DroAna_CAF1 34388 91 - 1 UUGGCAGGUGGCAGCGCUGCCGGCGUC-GCAGCAUUUGAAUAGCGCCGGCAGAGAAGCCGACUGCGACGUUGC----------CGCCGCCUAUGCAACCAAA ..(((.((((((((((((((((((((.-.(((...)))....))))))))))....((.....))..))))))----------)))))))............ ( -44.90) >DroPer_CAF1 31340 101 - 1 UUGGCAGGUGGCUGCGCUGCCGGCUUC-GCAGCAUUUGAAUAGCGCCAGCAUUGGCGCUGGCUUCGACGCCGGCGCCUGCCGCCGCCGCCUAUGCAACCAAA ..(((.((((((.((((.((((((.((-(.(((.......(((((((......)))))))))).))).))))))))..)).)))))))))............ ( -53.41) >consensus UUGGCAGGUGGCAGCGCUGCCGGCGUC_GCAGCAUUUGAAUAGCGCCAGCAGAGGCGCUGACUGCGACGCCGCCGCU______CGCCGCCUAUGCAACCAAA ...(((((((((.(((((((........)))))...((........))))...((((.((....)).)))).............))))))..)))....... (-27.66 = -26.88 + -0.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:23 2006