
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,417,720 – 24,417,814 |
| Length | 94 |
| Max. P | 0.863020 |

| Location | 24,417,720 – 24,417,814 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -22.49 |
| Energy contribution | -23.55 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24417720 94 + 27905053 ---------------GCUGACGUGCCAAGUGCCAUCGAAAAUGGCAAGUCAUUGGCAUUUCAACUGGACAUGCGGCACCCUUGUUAGCCCGAGCUGUCUCUGACUC----UUU ---------------(((((((((((...((((((.....))))))........((((.((.....)).)))))))))....))))))..((((.......).)))----... ( -29.20) >DroVir_CAF1 10581 98 + 1 ---GUUGACGCUGACGCUGACGGGCCAAGUGCCAUUGAAAAUGGCAAGUCAUUGGCUUUUCAACUGGACAUGCGGCACCCUUGUUAGCC---GCUGUCUGAGCC--------- ---......(((.....(((..((((((.((((((.....)))))).....))))))..)))...(((((.(((((..........)))---))))))).))).--------- ( -37.10) >DroPse_CAF1 14757 91 + 1 ---------------GCUGACGUGCCAAGUGCCAUUGAAAAUGGCAAGUCAUUGGCAUUUCAACUGGACAUGCGGCACCCUUGUUAGCCAGAG---UCUCUCUCUC----UCU ---------------(((((((((((..((((((((((((.((.(((....))).)))))))).))).)))..)))))....)))))).((((---.....)))).----... ( -32.30) >DroGri_CAF1 9946 101 + 1 GAGGCUGACGCUGACGCUGACGCGCCAAGUGCCAUUGAAAAUGGCAAGUCAUUUACAUUUCAACUGGACAUGCGGCACCCUUGUUAGUC---GCUGUCGGAGCC--------- ..((((((((((..(((....)))...)))(((((.....)))))..)))................((((.(((((..........)))---))))))..))))--------- ( -31.20) >DroMoj_CAF1 10576 101 + 1 GACGCUGACUCUGACGCUGACGUGCCAAGUACCAUUGAAAAUGGCAAGUCAUUGGCAUUUCAACUGGACAUGCGGCACCCUUGUUAACC---GCUGUCGUAGCC--------- ...((((..........(((.(((((((...((((.....)))).......))))))).)))....((((.((((............))---)))))).)))).--------- ( -27.20) >DroAna_CAF1 14072 98 + 1 ---------------GCUGACGUGCCAAGUGCUAUUGAAAAUGGCAAGUCAUUGGCAUUUCAACUGGACAUGCGGCACCCUUGAUAGCCCGAGCUGUCUCUGGUUCAGACCCA ---------------.((((.(((((..((((((((((((.((.(((....))).)))))))).))).)))..)))))((..((((((....))))))...)).))))..... ( -34.00) >consensus _______________GCUGACGUGCCAAGUGCCAUUGAAAAUGGCAAGUCAUUGGCAUUUCAACUGGACAUGCGGCACCCUUGUUAGCC___GCUGUCUCAGCC_________ ...............(((((((((((..((((((((((((.((.(((....))).)))))))).))).)))..)))))....))))))......................... (-22.49 = -23.55 + 1.06)



| Location | 24,417,720 – 24,417,814 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.719099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24417720 94 - 27905053 AAA----GAGUCAGAGACAGCUCGGGCUAACAAGGGUGCCGCAUGUCCAGUUGAAAUGCCAAUGACUUGCCAUUUUCGAUGGCACUUGGCACGUCAGC--------------- ...----......(.((((...((((((......))).)))..))))).(((((..((((((.(...(((((((...))))))))))))))..)))))--------------- ( -29.40) >DroVir_CAF1 10581 98 - 1 ---------GGCUCAGACAGC---GGCUAACAAGGGUGCCGCAUGUCCAGUUGAAAAGCCAAUGACUUGCCAUUUUCAAUGGCACUUGGCCCGUCAGCGUCAGCGUCAAC--- ---------(((.(.((((((---(((..........))))).))))..(((((...(((((.(...((((((.....))))))))))))...)))))....).)))...--- ( -35.90) >DroPse_CAF1 14757 91 - 1 AGA----GAGAGAGAGA---CUCUGGCUAACAAGGGUGCCGCAUGUCCAGUUGAAAUGCCAAUGACUUGCCAUUUUCAAUGGCACUUGGCACGUCAGC--------------- ...----......(.((---(..((((..........))))...)))).(((((..((((((.(...((((((.....)))))))))))))..)))))--------------- ( -26.90) >DroGri_CAF1 9946 101 - 1 ---------GGCUCCGACAGC---GACUAACAAGGGUGCCGCAUGUCCAGUUGAAAUGUAAAUGACUUGCCAUUUUCAAUGGCACUUGGCGCGUCAGCGUCAGCGUCAGCCUC ---------(((...((((((---((((......)))..))).))))...............((((.((((((.....)))))).(((((((....))))))).))))))).. ( -33.10) >DroMoj_CAF1 10576 101 - 1 ---------GGCUACGACAGC---GGUUAACAAGGGUGCCGCAUGUCCAGUUGAAAUGCCAAUGACUUGCCAUUUUCAAUGGUACUUGGCACGUCAGCGUCAGAGUCAGCGUC ---------((((..(((.((---(((..........))))).......(((((..((((((.(...((((((.....)))))))))))))..))))))))..))))...... ( -35.60) >DroAna_CAF1 14072 98 - 1 UGGGUCUGAACCAGAGACAGCUCGGGCUAUCAAGGGUGCCGCAUGUCCAGUUGAAAUGCCAAUGACUUGCCAUUUUCAAUAGCACUUGGCACGUCAGC--------------- ((((..((.(((...((.(((....))).))...)))....))..))))(((((..((((((((.((.............)))).))))))..)))))--------------- ( -26.92) >consensus _________GGCAGAGACAGC___GGCUAACAAGGGUGCCGCAUGUCCAGUUGAAAUGCCAAUGACUUGCCAUUUUCAAUGGCACUUGGCACGUCAGC_______________ ...............((((((...(((..........))))).))))..(((((..((((((.(...(((((((...))))))))))))))..)))))............... (-22.25 = -22.75 + 0.50)

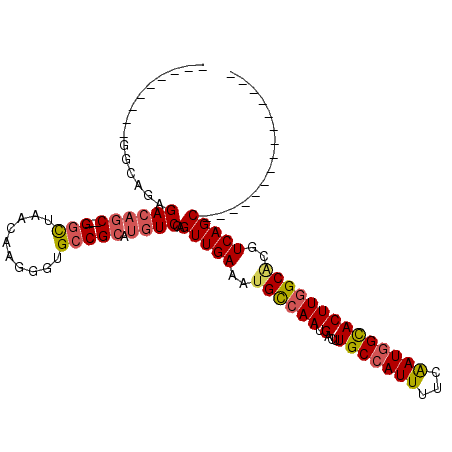
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:27:53 2006