
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,416,120 – 24,416,226 |
| Length | 106 |
| Max. P | 0.653534 |

| Location | 24,416,120 – 24,416,226 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -16.18 |
| Consensus MFE | -12.47 |
| Energy contribution | -12.67 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
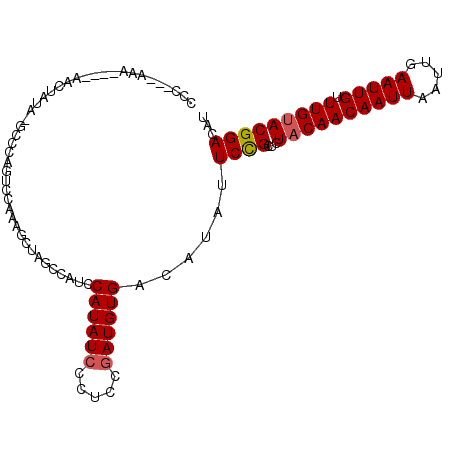
>3R_DroMel_CAF1 24416120 106 - 27905053 CCCCGAAAAAAAAAACUAUAUGCCCAGUCCAAAGCUAGCCAUCCAUAUCCCUCCGAUGUGACAUAUUCCGCUCUACAACAAUUAAUUGAAUUGUUUGUACGGACAU ..........................((((..(((........((((((.....)))))).........))).((((((((((.....))))).))))).)))).. ( -17.33) >DroSec_CAF1 9684 103 - 1 CUC---GAAAAAAAACUAUACGCCCAGUCCAAAGCUAGCCAUCCAUAUCCCUCCGAUGUGACAUAUUCCGCUCUACAACAAUUAAUUGAAUUGUUUGUACGGACAU ...---....................((((..(((........((((((.....)))))).........))).((((((((((.....))))).))))).)))).. ( -17.33) >DroSim_CAF1 7366 103 - 1 CUC---AAAAAAGAACUAUAUGCCCAGUCCAAAGCUAGCCAUCCAUAUCCCUCCGAUGUGACAUAUUCCGCUCUACAACAAUUAAUUGAAUUGUUUGUACGGACAU ...---....................((((..(((........((((((.....)))))).........))).((((((((((.....))))).))))).)))).. ( -17.33) >DroEre_CAF1 10440 99 - 1 CCU---AAA----UACUGUACGCCCAGUCCAAAGCCAGCCAUUCAUAUCCCACCGAUGUGACAUAUUCUGCUCUACAACAAUUAAUUGAAUUGUUUGUACGGACAU ...---...----.............((((..(((.((....(((((((.....)))))))......))))).((((((((((.....))))).))))).)))).. ( -19.50) >DroYak_CAF1 14212 86 - 1 CCU---AAA----UAC-------------CAAAGCUAGCCAUCCAUAUCCCUCCGAUGUGACAUAUUCCGCUCUACAACAAUUAAUUGAAUUGUUUGUACGGACAU ...---...----...-------------..............((((((.....))))))......((((...((((((((((.....))))).)))))))))... ( -14.20) >DroAna_CAF1 12500 88 - 1 CCC---AGC-------------UCCUUUCCGAAACUUACCAUCCAUAUCC--CCAAUGUGACAUAUUCCGCUCUACAACAAUUAAUUGAAUUGUUUGCACGGACAU ...---...-------------.....((((............(((((..--...))))).........((.....(((((((.....))))))).)).))))... ( -11.40) >consensus CCC___AAA____AACUAUA_GCCCAGUCCAAAGCUAGCCAUCCAUAUCCCUCCGAUGUGACAUAUUCCGCUCUACAACAAUUAAUUGAAUUGUUUGUACGGACAU ...........................................((((((.....))))))......((((...((((((((((.....))))).)))))))))... (-12.47 = -12.67 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:27:50 2006