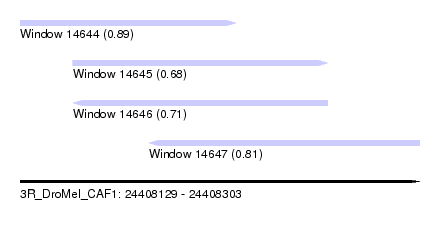
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,408,129 – 24,408,303 |
| Length | 174 |
| Max. P | 0.891311 |

| Location | 24,408,129 – 24,408,223 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -31.91 |
| Energy contribution | -31.98 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
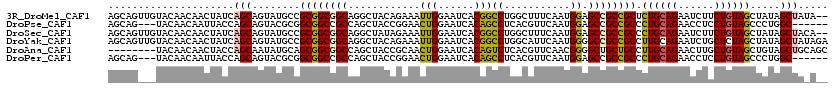
>3R_DroMel_CAF1 24408129 94 + 27905053 UGCACUCGCUAAUUGGCAGGCCCGCUACAAAGCCGCUGCCUGUGGAGAUCGGGAUC----UAUAGCUAUAGCUACAGAAGAUUCUGCAGAGCGGCGGC .((.((.(((....)))))))..........(((((((((((..(((.((....((----(.((((....)))).))).)))))..))).)))))))) ( -38.00) >DroSec_CAF1 1707 94 + 1 UGCACUCGCUAAUUGGCAGGCCCGCAACAAAGCCGCUGCCUGUGGAGAUCGGGAUC----UGUAGCUAUAGCUACAGAAGAUUCUGCAGGGCGGCGGC (((.(..(((....)))..)...))).....(((((((((((..(((.((....((----((((((....)))))))).)))))..)).))))))))) ( -44.10) >DroEre_CAF1 2397 94 + 1 UGCACUCGCUAAUUGGCUGGCCCGCAACAAAGCCGCUGCCUGUGGAGAUCGGGAUC----UAUAGCUAUAGCUAGAGCAGAUUCUGCAGGGCGGCGGC (((.((.(((....))).))...))).....(((((((((((..(((.((....((----(..(((....))))))...)))))..)).))))))))) ( -36.30) >DroYak_CAF1 5128 98 + 1 UGCACUCGCUAAUUGGCUGGCCCGCAACAAAGCCGCUGCCUGUGGAGAUCGGGAUCUAUCUAUAGCUAUAGCUAGAGCAGAUUCUGCAAGGCGGCGGC (((.((.(((....))).))...))).....(((((((((((..(((.((....((((.((((....)))).))))...)))))..).)))))))))) ( -38.10) >consensus UGCACUCGCUAAUUGGCAGGCCCGCAACAAAGCCGCUGCCUGUGGAGAUCGGGAUC____UAUAGCUAUAGCUACAGAAGAUUCUGCAGGGCGGCGGC .((.((.(((....))).))...))......((((((((((...(....(((((((......((((....)))).....)))))))).)))))))))) (-31.91 = -31.98 + 0.06)

| Location | 24,408,152 – 24,408,263 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -27.25 |
| Energy contribution | -28.78 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24408152 111 + 27905053 -GCUACAAAGCCGCUGCCUGUGGAGAUCGGGAUC----UAUAGCUAUAGCUACAGAAGAUUCUGCAGAGCGGCGGCUCCAUUGAAAGCCAGGCCGUGAUUCCAAUUUCUGUAGCCU -((((((.((((((((((((..(((.((....((----(.((((....)))).))).)))))..))).))))))))).....((((....((........))..)))))))))).. ( -44.00) >DroPse_CAF1 4700 100 + 1 UGCCACAAAGCCGCCGCUUGUG-----CGGG-----------GCCAGGGCUACAGGAGGUUCUGCAGGGCGGCGGCUCCAUUGAACGUGAGGCUGUGAUUCCAGUUCCGGUAGCUG ((((.((((((((((((((.((-----((((-----------(((..(....)....))))))))))))))))))))...))).....(..((((......))))..))))).... ( -44.70) >DroSec_CAF1 1730 111 + 1 -GCAACAAAGCCGCUGCCUGUGGAGAUCGGGAUC----UGUAGCUAUAGCUACAGAAGAUUCUGCAGGGCGGCGGCUCCAUUGAAAGCCAGGCCGUGAUUCCAAUUUCUAUAGCCU -.(((...((((((((((((..(((.((....((----((((((....)))))))).)))))..)).))))))))))...)))......((((..(((........)).)..)))) ( -47.60) >DroEre_CAF1 2420 111 + 1 -GCAACAAAGCCGCUGCCUGUGGAGAUCGGGAUC----UAUAGCUAUAGCUAGAGCAGAUUCUGCAGGGCGGCGGCCCCAUUGAAAGCCAGGCCGUGAUUCCAAUUUCUGUAGCCU -.(((....(((((((((((..(((.((....((----(..(((....))))))...)))))..)).)))))))))....)))......((((..(((........)).)..)))) ( -37.50) >DroYak_CAF1 5151 115 + 1 -GCAACAAAGCCGCUGCCUGUGGAGAUCGGGAUCUAUCUAUAGCUAUAGCUAGAGCAGAUUCUGCAAGGCGGCGGCCCCAUUGAAUGCCAGGCCGUGAUUCCAAUUUCUGUAGCCU -(((.(((.(((((((((((..(((.((....((((.((((....)))).))))...)))))..).))))))))))....)))..))).((((..(((........)).)..)))) ( -40.10) >DroPer_CAF1 4693 100 + 1 UGCCACAAAGCCGCCGCUUGUG-----CGGG-----------GCCAGGGCUACAGGAGGUUCUGCAGGGCGGCGGCUCCAUUGAACGUGAGGCUGUGAUUCCAGUUCCGGUAGCUG ((((.((((((((((((((.((-----((((-----------(((..(....)....))))))))))))))))))))...))).....(..((((......))))..))))).... ( -44.70) >consensus _GCAACAAAGCCGCUGCCUGUGGAGAUCGGGAUC____UAUAGCUAUAGCUACAGAAGAUUCUGCAGGGCGGCGGCUCCAUUGAAAGCCAGGCCGUGAUUCCAAUUUCUGUAGCCU .((..((((((((((((((.((.....(((((((......((((....)))).....))))))))))))))))))))...)))...))..(((...((........))....))). (-27.25 = -28.78 + 1.53)
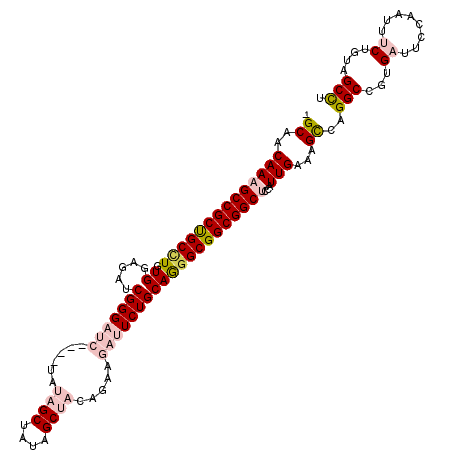
| Location | 24,408,152 – 24,408,263 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -24.95 |
| Energy contribution | -25.53 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24408152 111 - 27905053 AGGCUACAGAAAUUGGAAUCACGGCCUGGCUUUCAAUGGAGCCGCCGCUCUGCAGAAUCUUCUGUAGCUAUAGCUAUA----GAUCCCGAUCUCCACAGGCAGCGGCUUUGUAGC- ..((((((...((((((((((.....))).))))))).(((((((.((.(((..(((((.((((((((....))))))----))....))).))..))))).)))))))))))))- ( -42.20) >DroPse_CAF1 4700 100 - 1 CAGCUACCGGAACUGGAAUCACAGCCUCACGUUCAAUGGAGCCGCCGCCCUGCAGAACCUCCUGUAGCCCUGGC-----------CCCG-----CACAAGCGGCGGCUUUGUGGCA ..(((((((((.(((......)))..)).))......((((((((((((((((((......))))))....)))-----------...(-----(....)))))))))))))))). ( -39.60) >DroSec_CAF1 1730 111 - 1 AGGCUAUAGAAAUUGGAAUCACGGCCUGGCUUUCAAUGGAGCCGCCGCCCUGCAGAAUCUUCUGUAGCUAUAGCUACA----GAUCCCGAUCUCCACAGGCAGCGGCUUUGUUGC- ((((..(.((........)))..))))......(((..(((((((.(((.((..(((((.((((((((....))))))----))....))).))..))))).)))))))..))).- ( -44.30) >DroEre_CAF1 2420 111 - 1 AGGCUACAGAAAUUGGAAUCACGGCCUGGCUUUCAAUGGGGCCGCCGCCCUGCAGAAUCUGCUCUAGCUAUAGCUAUA----GAUCCCGAUCUCCACAGGCAGCGGCUUUGUUGC- (((((...((........))..)))))......((((..((((((.(((..((((...))))..((((....)))).(----(((....)))).....))).))))))..)))).- ( -35.80) >DroYak_CAF1 5151 115 - 1 AGGCUACAGAAAUUGGAAUCACGGCCUGGCAUUCAAUGGGGCCGCCGCCUUGCAGAAUCUGCUCUAGCUAUAGCUAUAGAUAGAUCCCGAUCUCCACAGGCAGCGGCUUUGUUGC- (((((...((........))..)))))......((((..((((((.((((((.((((((((((.((((....)))).)).))))).....))).)).)))).))))))..)))).- ( -40.90) >DroPer_CAF1 4693 100 - 1 CAGCUACCGGAACUGGAAUCACAGCCUCACGUUCAAUGGAGCCGCCGCCCUGCAGAACCUCCUGUAGCCCUGGC-----------CCCG-----CACAAGCGGCGGCUUUGUGGCA ..(((((((((.(((......)))..)).))......((((((((((((((((((......))))))....)))-----------...(-----(....)))))))))))))))). ( -39.60) >consensus AGGCUACAGAAAUUGGAAUCACGGCCUGGCUUUCAAUGGAGCCGCCGCCCUGCAGAAUCUCCUGUAGCUAUAGCUAUA____GAUCCCGAUCUCCACAGGCAGCGGCUUUGUUGC_ (((((...((........))..)))))........(..(((((((.(((.(((((......)))))((....))........................))).)))))))..).... (-24.95 = -25.53 + 0.58)

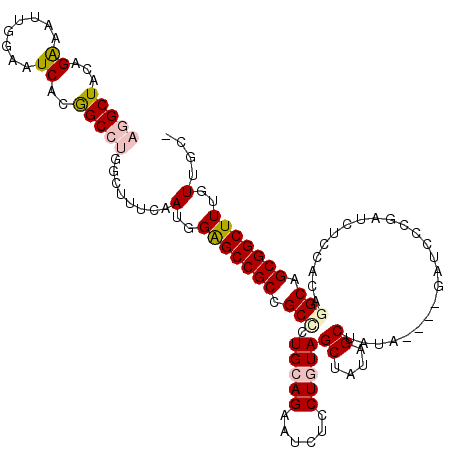
| Location | 24,408,185 – 24,408,303 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -40.65 |
| Consensus MFE | -24.67 |
| Energy contribution | -24.42 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24408185 118 - 27905053 AGCAGUUGUACAACAACUAUCAGCAGUAUGCCGCGGCGGCAGGCUACAGAAAUUGGAAUCACGGCCUGGCUUUCAAUGGAGCCGCCGCUCUGCAGAAUCUUCUGUAGCUAUAGCUAUA-- ...(((((.....)))))...(((....(((((...)))))(((((((((....(((.((..(((..(((((......))))))))((...)).)).))))))))))))...)))...-- ( -40.50) >DroPse_CAF1 4726 111 - 1 AGCAG---UACAACAAUUACCAGCAGUACGCGGCGGCCGCCAGCUACCGGAACUGGAAUCACAGCCUCACGUUCAAUGGAGCCGCCGCCCUGCAGAACCUCCUGUAGCCCUGGC------ .....---...........((((......((((((((..(((.....((((.(((......)))..)).)).....))).)))))))).((((((......))))))..)))).------ ( -38.50) >DroSec_CAF1 1763 118 - 1 AGCAGUUGUACAACAACUAUCAGCAGUAUGCCGCGGCGGCAGGCUAUAGAAAUUGGAAUCACGGCCUGGCUUUCAAUGGAGCCGCCGCCCUGCAGAAUCUUCUGUAGCUAUAGCUACA-- ...(((((.....)))))...((.((..(((.(.((((((((((..(.((........)))..))))(((((......)))))))))))).)))....)).))(((((....))))).-- ( -43.00) >DroYak_CAF1 5186 120 - 1 AGCAGUUGUACAACAACUAUCAGCAGUAUGCCGCGGCGGCAGGCUACAGAAAUUGGAAUCACGGCCUGGCAUUCAAUGGGGCCGCCGCCUUGCAGAAUCUGCUCUAGCUAUAGCUAUAGA ...(((((.....)))))...(((((..(((.(((((((((((((...((........))..)))))..(((...)))..))))))))...)))....))))).((((....)))).... ( -40.40) >DroAna_CAF1 4740 112 - 1 --------UACAACAACUACCAGCAAUAUGCAGCGGCGGCCAGCUACCGCAACUGGAAUCACAGUCUCACGUUCAACGGGGCUGCUGCCUUGCAGAACUUGCUGUAGCUGUAGCUGCAGC --------..............((.....)).(((((...(((((((.(((((((......)))).....((((..((((((....))))))..)))).))).)))))))..)))))... ( -43.00) >DroPer_CAF1 4719 111 - 1 AGCAG---UACAACAAUUACCAGCAGUACGCGGCGGCCGCCAGCUACCGGAACUGGAAUCACAGCCUCACGUUCAAUGGAGCCGCCGCCCUGCAGAACCUCCUGUAGCCCUGGC------ .....---...........((((......((((((((..(((.....((((.(((......)))..)).)).....))).)))))))).((((((......))))))..)))).------ ( -38.50) >consensus AGCAG___UACAACAACUACCAGCAGUAUGCCGCGGCGGCAAGCUACAGAAACUGGAAUCACAGCCUCACGUUCAAUGGAGCCGCCGCCCUGCAGAACCUCCUGUAGCUAUAGCUA_A__ .....................(((........((((((((............(((......)))((...........)).)))))))).((((((......)))))).....)))..... (-24.67 = -24.42 + -0.25)
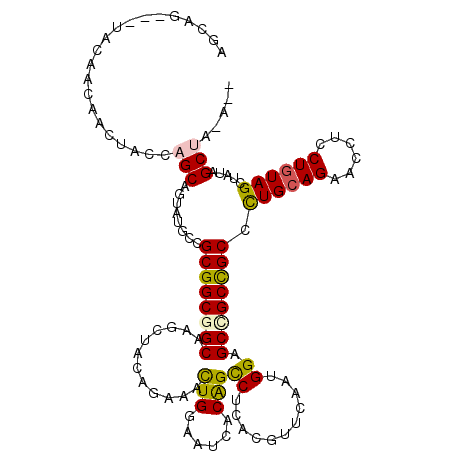
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:27:47 2006