
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,403,686 – 24,403,793 |
| Length | 107 |
| Max. P | 0.971952 |

| Location | 24,403,686 – 24,403,793 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 94.41 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -17.03 |
| Energy contribution | -16.59 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
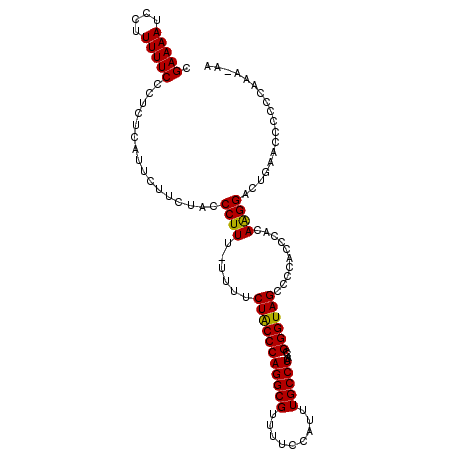
>3R_DroMel_CAF1 24403686 107 + 27905053 CGAAAAUCCUUUUUCCCUCUCAUUCUUCUACCCUUUUUUUUCUGCCCAGGCGUUUUCCAUUUGCCUAACCGGGUAGCCCCACCCACAAGGACUAAACCCCCCGAA-AA .(((((....)))))................((((......(((((((((((.........)))))....))))))..........))))...............-.. ( -16.69) >DroSec_CAF1 225765 105 + 1 CGAAAAUCCUUUUUCCCUCUCAUUCUUCUACCCUUU-UUUUCUACCCAGGCGUUUUCCAUUUGCCUAACCGGGUAGCCCCACCCACAGGGACUGAACCCUCCAAA--A .(((((....))))).....................-....(((((((((((.........)))))....))))))..........((((......)))).....--. ( -19.50) >DroSim_CAF1 234997 107 + 1 CGAAAAUCCUUUUUCCCUCUCAUGCUUCUACCCUUU-UUUUCUACCCAGGCGUUUUCCAUUUGCCUAACCGGGUAGCCCCACCCACAAGGACUGAACCCCCCAAAAAA .(((((....)))))....(((.(.((((.......-....(((((((((((.........)))))....))))))...........))))))))............. ( -17.15) >consensus CGAAAAUCCUUUUUCCCUCUCAUUCUUCUACCCUUU_UUUUCUACCCAGGCGUUUUCCAUUUGCCUAACCGGGUAGCCCCACCCACAAGGACUGAACCCCCCAAA_AA .(((((....)))))................((((......(((((((((((.........)))))....))))))..........)))).................. (-17.03 = -16.59 + -0.44)


| Location | 24,403,686 – 24,403,793 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.41 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -28.21 |
| Energy contribution | -27.33 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
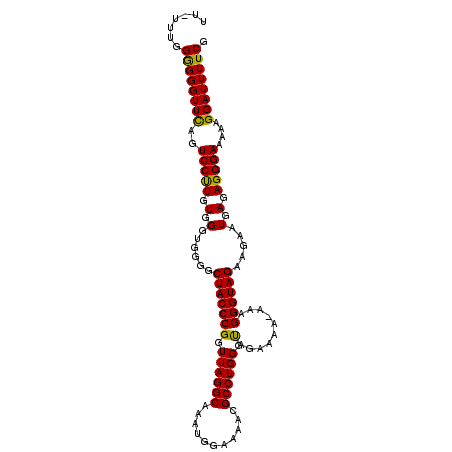
>3R_DroMel_CAF1 24403686 107 - 27905053 UU-UUCGGGGGGUUUAGUCCUUGUGGGUGGGGCUACCCGGUUAGGCAAAUGGAAAACGCCUGGGCAGAAAAAAAAGGGUAGAAGAAUGAGAGGGAAAAAGGAUUUUCG ..-....(..(((((..(((((.(.(......(((((((.((((((...........)))))).)..........)))))).....).).)))))....)))))..). ( -26.70) >DroSec_CAF1 225765 105 - 1 U--UUUGGAGGGUUCAGUCCCUGUGGGUGGGGCUACCCGGUUAGGCAAAUGGAAAACGCCUGGGUAGAAAA-AAAGGGUAGAAGAAUGAGAGGGAAAAAGGAUUUUCG .--....((((((((..(((((.(.(......(((((((.((((((...........)))))).)......-...)))))).....).).)))))....)))))))). ( -30.10) >DroSim_CAF1 234997 107 - 1 UUUUUUGGGGGGUUCAGUCCUUGUGGGUGGGGCUACCCGGUUAGGCAAAUGGAAAACGCCUGGGUAGAAAA-AAAGGGUAGAAGCAUGAGAGGGAAAAAGGAUUUUCG .......(..(((((..(((((.(.(((....(((((((.((((((...........)))))).)......-...))))))..)).).).)))))....)))))..). ( -28.30) >consensus UU_UUUGGGGGGUUCAGUCCUUGUGGGUGGGGCUACCCGGUUAGGCAAAUGGAAAACGCCUGGGUAGAAAA_AAAGGGUAGAAGAAUGAGAGGGAAAAAGGAUUUUCG .......((((((((..(((((.(.(......(((((((.((((((...........)))))).)..........)))))).....).).)))))....)))))))). (-28.21 = -27.33 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:27:43 2006