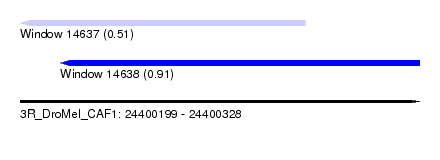
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,400,199 – 24,400,328 |
| Length | 129 |
| Max. P | 0.911723 |

| Location | 24,400,199 – 24,400,291 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -16.90 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.02 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24400199 92 - 27905053 AUCUUUAGCCAAUAAAGAAAACAGGAUAAGCCAUCAUUUCCGAAUUGCAAGUCAAAAGGAUGUAAGCAGGCGGAAUUCCCCGCAA------------------AACAUCA .((((((.....))))))......(((..((..((......))...))..))).....(((((......((((......))))..------------------.))))). ( -17.30) >DroVir_CAF1 214506 110 - 1 AUCUUUAACCAAUAAAGAAAACAGGAUAAGCCAGCAUUUCCGAAUUGCAAGUCAAAAAGAUUUAAGUAGGCGGAAUUCCCCACAACCAAAUAAUAAUGCAAACAACAACA .((((((.....)))))).....((....(((.(((.........)))(((((.....))))).....)))((......))....))....................... ( -16.20) >DroEre_CAF1 236763 92 - 1 AUCUUUAGCCAAUAAAGAAAACAGGAUAAGCCAUCAUUUCCGAUUUGCAAGUCAAAAGGAUGUAAGCAGGCGGAAUUCCCCGCAA------------------AACAUCA .((((((.....))))))......(((..((.(((......)))..))..))).....(((((......((((......))))..------------------.))))). ( -18.40) >DroWil_CAF1 206062 95 - 1 -UCUUUAGCCAAUAAAGAAAACAGGAUAAUCCACUAUUUCCGAAUUGCAAGUCAAAAAGAUUUAAGCAAGCGGAAUUCCCCGCAAUUGA--------------AGAAUGA -(((((((...............(((............)))...(((((((((.....)))))..))))((((......))))..))))--------------))).... ( -17.90) >DroYak_CAF1 230723 92 - 1 AUCUUAAGCCAAUAAAGAAAACAGGAUAAGCCAUCAUUUCCGAAUUGCAAGUCAAAAGGAUGUAAGCAGGCGGAAUUCCCCGCAA------------------AACAUCA .......((.(((...((((...((.....))....))))...)))))..........(((((......((((......))))..------------------.))))). ( -13.90) >DroAna_CAF1 225908 91 - 1 AUCUUUAACCAAUAAAGAAAACAGGAUAAGCCAUCAUUUCCGAAUUGCAAGUCAAAAAGAUGUAAGCAAGCGGAAUUC-CCGCAG------------------CACAUCA .((((((.....))))))......(((..((..((......))...))..))).....(((((..((..((((.....-)))).)------------------)))))). ( -17.70) >consensus AUCUUUAGCCAAUAAAGAAAACAGGAUAAGCCAUCAUUUCCGAAUUGCAAGUCAAAAAGAUGUAAGCAGGCGGAAUUCCCCGCAA__________________AACAUCA .((((((.....)))))).....(((............)))...((((..(((.....)))....))))((((......))))........................... (-14.04 = -14.02 + -0.03)



| Location | 24,400,212 – 24,400,328 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.58 |
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24400212 116 - 27905053 AAAACAAGGACCAUUCAAAGCAGCGACGACUCACGGCAUCUUUAGCCAAUAAAGAAAACAGGAUAAGCCAUCAUUUCCGAAUUGCAAGUCAAA-AGGAUGUAAGCAGGCGGAAUUCC .......((...((((...((.(.((....)).).)).((((((.....)))))).....))))...)).....(((((..((((..(((...-..)))....)))).))))).... ( -20.70) >DroVir_CAF1 214537 109 - 1 AAAACAAAGCGUACAUUAGGCAACAA-------CAGAAUCUUUAACCAAUAAAGAAAACAGGAUAAGCCAGCAUUUCCGAAUUGCAAGUCAAA-AAGAUUUAAGUAGGCGGAAUUCC ........((.(((.....((((...-------.....((((((.....)))))).....(((...(....)...)))...))))(((((...-..)))))..))).))((....)) ( -18.00) >DroGri_CAF1 209952 109 - 1 AAAACAAAGCAAACAUUAAGCAACAA-------CAGAAUCUUUAACCAAUAAAGAAAACAGGAUAAGCCAGUAUUUCCGAAUUGCAAGUCAAA-AAGAUUUAAGUAGGCGGAAUUCC ........((..((.....((((...-------.....((((((.....)))))).....(((............)))...))))(((((...-..)))))..))..))((....)) ( -14.50) >DroWil_CAF1 206079 92 - 1 ACAACAAAG-CCAUU-----------------------UCUUUAGCCAAUAAAGAAAACAGGAUAAUCCACUAUUUCCGAAUUGCAAGUCAAA-AAGAUUUAAGCAAGCGGAAUUCC .........-...((-----------------------((((((.....))))))))...((.....)).....(((((..(((((((((...-..)))))..)))).))))).... ( -18.30) >DroMoj_CAF1 190466 110 - 1 AAAACAAAGCGUACAUUAAGCAACAA-------CAGAUUCUUUAACCAAUAAAGAAAAUAGGAUAAGCCAGAGUUUCCGAAUUGCAAGUCAAAAAAGACUUAAGUAGGCGGAAUUCC ........((.(((.....((((...-------....(((((((.....)))))))....(((..(((....))))))...))))(((((......)))))..))).))((....)) ( -21.50) >DroAna_CAF1 225921 111 - 1 AAAACAAAAACAUUGCAGACC---G-AGACACACUGCAUCUUUAACCAAUAAAGAAAACAGGAUAAGCCAUCAUUUCCGAAUUGCAAGUCAAA-AAGAUGUAAGCAAGCGGAAUUC- .............(((((...---.-.......)))))((((((.....)))))).....((.....)).....(((((..((((..(((...-..)))....)))).)))))...- ( -18.60) >consensus AAAACAAAGCCAACAUUAAGCAACAA_______CAGAAUCUUUAACCAAUAAAGAAAACAGGAUAAGCCAGCAUUUCCGAAUUGCAAGUCAAA_AAGAUUUAAGCAGGCGGAAUUCC ......................................((((((.....)))))).....((.....)).....(((((..(((((((((......)))))..)))).))))).... (-14.38 = -14.10 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:27:40 2006