| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,397,227 – 24,397,318 |
| Length | 91 |
| Max. P | 0.873337 |

| Location | 24,397,227 – 24,397,318 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -19.14 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.16 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
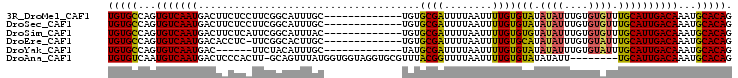
>3R_DroMel_CAF1 24397227 91 + 27905053 CUGUGCAUUUGUCAAUGCAAACACACAAAUAUAUACACAAAAUUAAAAUCGCACA-------------GCAAAUGCCGAAGGAGAAGUCAUUGACACUGGCACA ..((((...(((((((((................................(((..-------------.....)))(....)....).))))))))...)))). ( -18.30) >DroSec_CAF1 219317 91 + 1 CUGUGCAUUUGUCAAUGCAAACACACAAAUAUAUACACAAAAUUAAAAUCGCACA-------------GCAAAUGCCGAAGGAGAAGUCAUUGACACUGGCACA ..((((...(((((((((................................(((..-------------.....)))(....)....).))))))))...)))). ( -18.30) >DroSim_CAF1 227995 91 + 1 CUGUGCAUUUGUCAAUGCAAACACACAAAUAUACACACAAAAUUAAAAUCGCACA-------------GUAAAUGCCGAAUGAGAAGUCAUUGACACUGGCACA ..((((...(((((((((................................(((..-------------.....)))(......)..).))))))))...)))). ( -16.60) >DroEre_CAF1 233889 90 + 1 CUGUGCAUUUGUCAAUGCAAAUACACAAAUAUAUGCACAAAAUUAAAAUCGCACA-------------GCAAGUGCCGAA-GAGGUGUCAUUGACACUGGCACA ((((((((((.....((((.(((......))).))))........)))).)))))-------------)...(((((...-..((((((...))))))))))). ( -25.62) >DroYak_CAF1 227779 85 + 1 CUGUGCAUUUGUCAAUGCAAAUACACAAAUAUAUACACAAAAUUAAAAUCGCAUA-------------GCAAAUGUAGAA------GUCAUUGACACUGGCACA ..((((...(((((((((...((((......(((.(..............).)))-------------.....))))...------).))))))))...)))). ( -16.84) >DroAna_CAF1 222959 95 + 1 CUGUGCAUUUGUCAAUGCA--------AAUAUAUACACAAAAUUAAAACCGUAAACGCACCUACCACCAUAAACUGC-AAGUGGGAGUCAUUGACAUUGACACA ..(((((..(((((((((.--------......................((....))..(((((...((.....)).-..))))).).)))))))).)).))). ( -19.20) >consensus CUGUGCAUUUGUCAAUGCAAACACACAAAUAUAUACACAAAAUUAAAAUCGCACA_____________GCAAAUGCCGAAGGAGAAGUCAUUGACACUGGCACA ..((((...(((((((((................................((................))......(......)..).))))))))...)))). (-12.32 = -12.16 + -0.17)

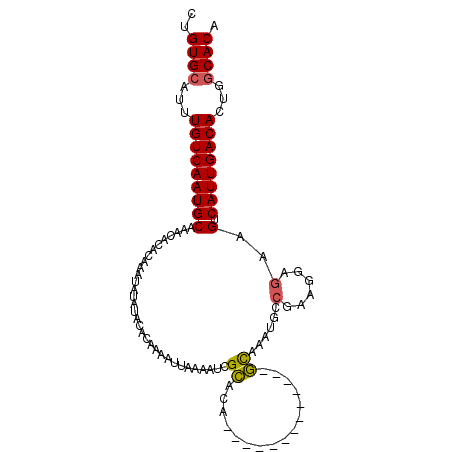
| Location | 24,397,227 – 24,397,318 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -17.84 |
| Energy contribution | -17.65 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24397227 91 - 27905053 UGUGCCAGUGUCAAUGACUUCUCCUUCGGCAUUUGC-------------UGUGCGAUUUUAAUUUUGUGUAUAUAUUUGUGUGUUUGCAUUGACAAAUGCACAG (((((...((((((((.....((...((((....))-------------))...))..........(..((((....))))..)...))))))))...))))). ( -23.30) >DroSec_CAF1 219317 91 - 1 UGUGCCAGUGUCAAUGACUUCUCCUUCGGCAUUUGC-------------UGUGCGAUUUUAAUUUUGUGUAUAUAUUUGUGUGUUUGCAUUGACAAAUGCACAG (((((...((((((((.....((...((((....))-------------))...))..........(..((((....))))..)...))))))))...))))). ( -23.30) >DroSim_CAF1 227995 91 - 1 UGUGCCAGUGUCAAUGACUUCUCAUUCGGCAUUUAC-------------UGUGCGAUUUUAAUUUUGUGUGUAUAUUUGUGUGUUUGCAUUGACAAAUGCACAG (((((...((((((((.(....(((.(((.((.(((-------------.(..(((........)))..)))).))))).)))...)))))))))...))))). ( -23.90) >DroEre_CAF1 233889 90 - 1 UGUGCCAGUGUCAAUGACACCUC-UUCGGCACUUGC-------------UGUGCGAUUUUAAUUUUGUGCAUAUAUUUGUGUAUUUGCAUUGACAAAUGCACAG (((((...((((((((.((....-....((((....-------------.))))............(((((((....))))))).))))))))))...))))). ( -29.20) >DroYak_CAF1 227779 85 - 1 UGUGCCAGUGUCAAUGAC------UUCUACAUUUGC-------------UAUGCGAUUUUAAUUUUGUGUAUAUAUUUGUGUAUUUGCAUUGACAAAUGCACAG (((((...((((((((.(------...(((((....-------------(((((((........))))))).......)))))...)))))))))...))))). ( -24.30) >DroAna_CAF1 222959 95 - 1 UGUGUCAAUGUCAAUGACUCCCACUU-GCAGUUUAUGGUGGUAGGUGCGUUUACGGUUUUAAUUUUGUGUAUAUAUU--------UGCAUUGACAAAUGCACAG (((((...((((((((....(((((.-.........)))))((((((..(.(((((........))))).)..))))--------))))))))))...))))). ( -23.80) >consensus UGUGCCAGUGUCAAUGACUUCUCCUUCGGCAUUUGC_____________UGUGCGAUUUUAAUUUUGUGUAUAUAUUUGUGUGUUUGCAUUGACAAAUGCACAG (((((...(((((((.....................................((((........))))(((.((((....)))).))))))))))...))))). (-17.84 = -17.65 + -0.19)
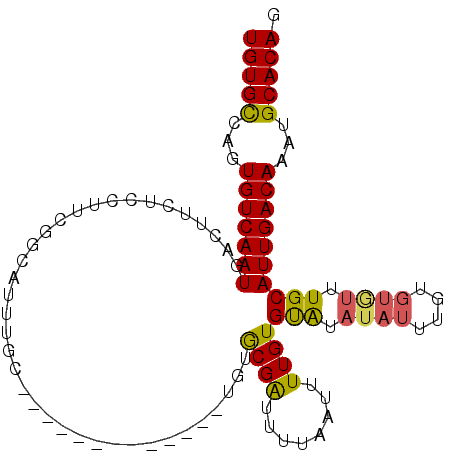
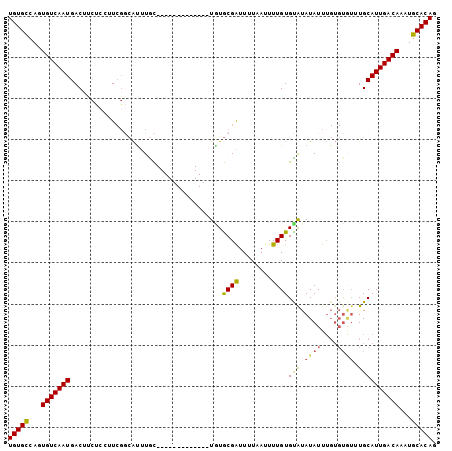
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:27:36 2006