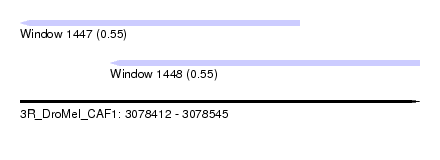
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,078,412 – 3,078,545 |
| Length | 133 |
| Max. P | 0.552825 |

| Location | 3,078,412 – 3,078,505 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -18.38 |
| Consensus MFE | -12.25 |
| Energy contribution | -12.17 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
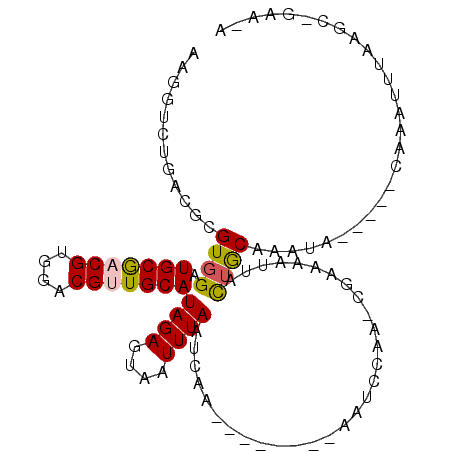
>3R_DroMel_CAF1 3078412 93 - 27905053 AAGGUCUGACGCGUGGAUGCGACGUGGACGUUGCAUAGAGUAAUUUAAUCAG--------AAUCCAAGCGAAAAUUACAGCGAGUA-----AAAUUUUAAGC-GAA-A ...(..((((((.(((((((((((....)))))).((((....)))).....--------.))))).)))....((((.....)))-----)....)))..)-...-. ( -22.30) >DroVir_CAF1 4391 87 - 1 AAGGUCUGACGUGUGGAUGCAACGUGGACGCUGCAUAGAGUAAUUUAAUGAA--------AAUUCCAUCAAGAAUUACUGCA-----------AAAUUAAGC-GAA-A ...((((.((((((....)).))))))))(((.....(((((((((..(((.--------.......))).)))))))).).-----------......)))-...-. ( -16.20) >DroPse_CAF1 4073 93 - 1 AAGGUCUGACGCGUGGAUGCGACGUGGACGUUGCAUAGAGUAAUUUAAUCAA--------AUUCCAA-CGAAAAUUACUACAAAUU-----CAAAUUUAAGC-GAAAA .........(((....((((((((....))))))))..((((((((..((..--------.......-.)))))))))).......-----.........))-).... ( -19.10) >DroGri_CAF1 4423 104 - 1 AAGGUCUGACGUGUGGAUGCAACGUGGACGCUGCAUAGAGUAAUUUAAA---ACAAAAACAAUUCCAUCAAAAAUUACUGCAAGAAAGCUACAAAAUUAAGAACAA-A ..(.(((....((((((((((.((....)).)))))..((((((((...---...................)))))))).........)))))......))).)..-. ( -16.75) >DroAna_CAF1 4488 91 - 1 AAGGUCUGACGCGUGGAUGCGACGUGGACGUUGCAUAGAGUAAUUUAAUCAA--------AAUC--AGCGAAAAUGAUUGCAAAUA-----AAAUUUUAAAC-GAA-A ..((((.....(((.(((((((((....)))))).((((....)))).....--------.)))--.))).....)))).......-----...........-...-. ( -16.80) >DroPer_CAF1 4085 93 - 1 AAGGUCUGACGCGUGGAUGCGACGUGGACGUUGCAUAGAGUAAUUUAAUCAA--------AUUCCAA-CGAAAAUUACUACAAAUU-----CAAAUUUAAGC-GAAAA .........(((....((((((((....))))))))..((((((((..((..--------.......-.)))))))))).......-----.........))-).... ( -19.10) >consensus AAGGUCUGACGCGUGGAUGCGACGUGGACGUUGCAUAGAGUAAUUUAAUCAA________AAUCCAA_CGAAAAUUACUGCAAAUA_____CAAAUUUAAGC_GAA_A ............((((.(((((((....)))))))((((....))))..............................))))........................... (-12.25 = -12.17 + -0.08)

| Location | 3,078,442 – 3,078,545 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
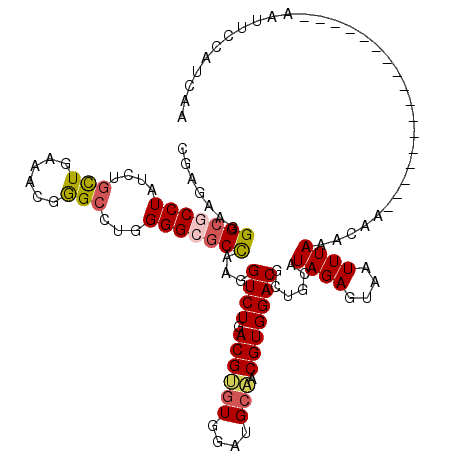
>3R_DroMel_CAF1 3078442 103 - 27905053 UGAGAAGGGCGCCUACCUGCUGAAGAGGGCCUGGGGCGCCAAGGUCUGACGCGUGGAUGCGACGUGGACGUUGCAUAGAGUAAUUUAAUCAG-----------------AAUCCAAGCGA ..(((..(((((((....(((......)))...)))))))....)))..(((.(((((((((((....)))))).((((....)))).....-----------------.))))).))). ( -40.20) >DroVir_CAF1 4415 103 - 1 GGAGAAGGGCGCCUAUCUAUUGAAACGGGCCUGGGGCGCCAAGGUCUGACGUGUGGAUGCAACGUGGACGCUGCAUAGAGUAAUUUAAUGAA-----------------AAUUCCAUCAA ((((...((((((............(((((((((....)).)))))))((((((....)).)))))).)))).((((((....))).)))..-----------------..))))..... ( -28.10) >DroGri_CAF1 4459 108 - 1 CGAGAAGGGAGCCUAUCUGCUGAAACGAGCCUGGGGCGCCAAGGUCUGACGUGUGGAUGCAACGUGGACGCUGCAUAGAGUAAUUUAAA------------ACAAAAACAAUUCCAUCAA ..(((..((.((((....(((......)))...)))).))....))).....(((((((((.((....)).))))((((....))))..------------...........)))))... ( -25.40) >DroWil_CAF1 11851 103 - 1 AGAGAAGGGCUCCUAUCUGCUCAAGAGAGCCUGGGGUGCUAAGGUCUGACGAUUGGAUGCAACGUGGACACUGCAUAGAGUAAUUUAAAUGA-----------------AAAAAAAACAA ...(((..((((((....((((....))))...))((((....((((.(((.(((....))))))))))...)))).))))..)))......-----------------........... ( -23.10) >DroMoj_CAF1 4611 120 - 1 CGAAAAGGGCGCCUAUCUGCUGAAACGGGCCUGGGGAGCCAAGGUCUGACGUGUGGAUGCAACGUGGACGCUGCAUAGAGUAAUUUAAAAGAAUAAACAAAACAAAACCAAUUCCAUGAA .......((((......))))....(((((((((....)).)))))))....(((((((((.((....)).))))((((....)))).........................)))))... ( -27.40) >DroAna_CAF1 4518 101 - 1 CGAGCGGGGCGCCUACCUGCUGAAGCGGGCAUGGGGCGCCAAGGUCUGACGCGUGGAUGCGACGUGGACGUUGCAUAGAGUAAUUUAAUCAA-----------------AAUC--AGCGA .((.(..(((((((.(((((....)))))....)))))))..).))...(((...(((((((((....)))))).((((....)))).....-----------------.)))--.))). ( -42.70) >consensus CGAGAAGGGCGCCUAUCUGCUGAAACGGGCCUGGGGCGCCAAGGUCUGACGUGUGGAUGCAACGUGGACGCUGCAUAGAGUAAUUUAAACAA_________________AAUUCCAUCAA .......(((((((....(((......)))...)))))))...((((.((((((....))).)))))))......((((....))))................................. (-21.95 = -22.12 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:16 2006