| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,383,233 – 24,383,457 |
| Length | 224 |
| Max. P | 0.953314 |

| Location | 24,383,233 – 24,383,340 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -21.21 |
| Consensus MFE | -14.75 |
| Energy contribution | -15.87 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
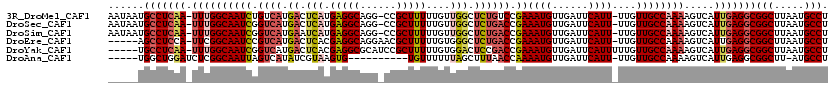
>3R_DroMel_CAF1 24383233 107 - 27905053 ACUCGCGGAUUAACGCUAGUGCACAUAUAUUGAAAAC-AAAAUGACACACAGUAGGCCUCGUUCCCUAUGCAAAGGACGAUUUAAGUCAUAAGCUCAGCAACAAUAUU (((.(((......))).)))......((((((.....-.............(((((........)))))((....(((.......)))....)).......)))))). ( -17.50) >DroSec_CAF1 205482 98 - 1 ACUCGCGGAUUAACGCUAGUGCACAUUUAUUCAAAACCAUAAUGGCACACAAAGGAAGAGGUCCUUUA----------AAUUUGAGUCAUAAGCUCAGAAACAAUAUU ....(((......)))..((((.((((.............))))))))..((((((.....)))))).----------..(((((((.....)))))))......... ( -22.52) >DroSim_CAF1 214101 98 - 1 ACUCGCGGAUUAACGCUAGUGCACAUUUAUUAAAACCCAGAAUGGCACACAAAGGAAGAGGUCCUUUA----------AAUUUGAGUCAAAAGCUCAAAAACAAUAUU ....(((......)))..((((.(((((...........)))))))))..((((((.....)))))).----------..(((((((.....)))))))......... ( -23.60) >consensus ACUCGCGGAUUAACGCUAGUGCACAUUUAUUAAAAACCAAAAUGGCACACAAAGGAAGAGGUCCUUUA__________AAUUUGAGUCAUAAGCUCAGAAACAAUAUU ....(((......)))..((((.(((((...........)))))))))..((((((.....)))))).............(((((((.....)))))))......... (-14.75 = -15.87 + 1.12)


| Location | 24,383,340 – 24,383,457 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -26.94 |
| Energy contribution | -27.87 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24383340 117 + 27905053 AAUAAUGCCUCAA-UUUGGCAAUCUGUCAUGACUCAUGAGGCAGG-CCGCUUUUUGUUGGCUCUGUCCGAAAUGUUGAUUCAUU-UUGUUGCCAAAAGUCAUUGAGGCGGCUUAAUGCCU ......(((((((-((((((((....(((((...)))))((((((-((((.....).)))).)))))(((((((......))))-)))))))))).....))))))))(((.....))). ( -43.70) >DroSec_CAF1 205580 117 + 1 AAUAAUGCCUCAA-UUUGGCAAUCGGUCAUGACUCAUGAGGCAGG-CCGCUUUUUGUUGGCUCUGACCGAAAUGUUGAUUCAUU-UUGUUGCCAAAAGUCAUUGAGGCGGCUUAAUGCCU ......(((((((-(((((((((.(((((.((..((.(((((...-..)))))....))..)))))))((((((......))))-)))))))))).....))))))))(((.....))). ( -43.50) >DroSim_CAF1 214199 117 + 1 AAUAAUGCCUCAA-UUUGGCAAUCGGUCAUGAAUCAUGAGGCAGG-CCGCUUUUUGUUGGCUCUGACCGAAAUGUUGAUUCAUU-UUGUUGCCAAAAGUCAUUGAGGCGGCUUAAUGCCU ......(((((((-((((((((.(((..((((((((...((((((-((((.....).)))).))).)).......)))))))).-)))))))))).....))))))))(((.....))). ( -44.90) >DroEre_CAF1 219773 113 + 1 -----AGCCUCCA-UUCGGCAAUCCGUCAUGACUCACGAGGCAGGAACGCUUUUUGUGGGCUCUGACCGAAAUGUUGAUUCAUU-UUGUUGCCAAAAGUCAUUGAGGCGGCUUAAUGCCU -----.(((((..-...(((((...((((.((((((((((((......)))..))))))).))))))(((((((......))))-))))))))..((....)))))))(((.....))). ( -38.20) >DroYak_CAF1 213304 114 + 1 -----UGCCUCAA-UUUGGCAAUCGGUCAUGACUCACGAGGCGCAUCCGCUUUUUGUGGACUCCGACCGAAAUGUUGAUUCAUUUUUGUUGCCAAAAGUCAUUGAGGCGGCUUAAUGCCU -----.(((((((-((((((((((((((.....((((((((.((....)).)))))))).....)))))(((((......)))))..)))))))).....))))))))(((.....))). ( -43.10) >DroAna_CAF1 208314 103 + 1 -----UGGCUGGAUCUCGGCAAUUAGUCAUAUCGUAAGUG----------UGUUUUUUAGCUUUAACCAAAAUGUUGAUUCAUU-UUGUUGCCAAAAGUCAUUGAGGCGGCUU-AUGCCU -----.(((........(((((.....((((.(....)))----------))...............(((((((......))))-))))))))..(((((........)))))-..))). ( -22.50) >consensus _____UGCCUCAA_UUUGGCAAUCGGUCAUGACUCAUGAGGCAGG_CCGCUUUUUGUUGGCUCUGACCGAAAUGUUGAUUCAUU_UUGUUGCCAAAAGUCAUUGAGGCGGCUUAAUGCCU ......(((((((.((((((((((.((((.((.(((.(((((......)))))....))).)))))).))((((......))))....)))))))).....)))))))(((.....))). (-26.94 = -27.87 + 0.92)


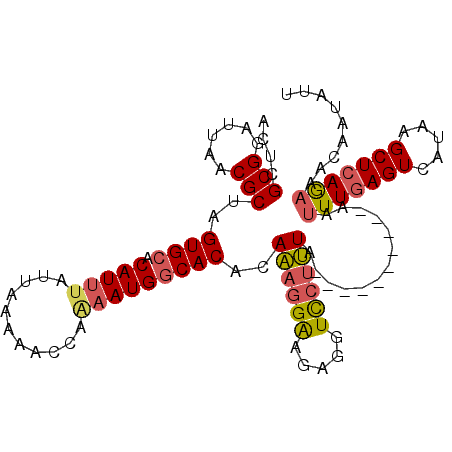
| Location | 24,383,340 – 24,383,457 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -28.57 |
| Energy contribution | -28.75 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24383340 117 - 27905053 AGGCAUUAAGCCGCCUCAAUGACUUUUGGCAACAA-AAUGAAUCAACAUUUCGGACAGAGCCAACAAAAAGCGG-CCUGCCUCAUGAGUCAUGACAGAUUGCCAAA-UUGAGGCAUUAUU .(((.....)))((((((((.....((((((((.(-((((......))))).((.(((.(((..........))-))))))(((((...)))))..).))))))))-)))))))...... ( -37.30) >DroSec_CAF1 205580 117 - 1 AGGCAUUAAGCCGCCUCAAUGACUUUUGGCAACAA-AAUGAAUCAACAUUUCGGUCAGAGCCAACAAAAAGCGG-CCUGCCUCAUGAGUCAUGACCGAUUGCCAAA-UUGAGGCAUUAUU .(((.....)))((((((((.....(((((((...-((((......))))(((((((..(((..........))-).((.((....)).)))))))))))))))))-)))))))...... ( -38.30) >DroSim_CAF1 214199 117 - 1 AGGCAUUAAGCCGCCUCAAUGACUUUUGGCAACAA-AAUGAAUCAACAUUUCGGUCAGAGCCAACAAAAAGCGG-CCUGCCUCAUGAUUCAUGACCGAUUGCCAAA-UUGAGGCAUUAUU .(((.....)))((((((((.....(((((((...-.((((((((.......((.(((.(((..........))-))))))...))))))))......))))))))-)))))))...... ( -40.90) >DroEre_CAF1 219773 113 - 1 AGGCAUUAAGCCGCCUCAAUGACUUUUGGCAACAA-AAUGAAUCAACAUUUCGGUCAGAGCCCACAAAAAGCGUUCCUGCCUCGUGAGUCAUGACGGAUUGCCGAA-UGGAGGCU----- .(((.....)))(((((.......((((((((...-((((......))))((.((((..((.(((.....(((....)))...))).))..)))).))))))))))-..))))).----- ( -34.50) >DroYak_CAF1 213304 114 - 1 AGGCAUUAAGCCGCCUCAAUGACUUUUGGCAACAAAAAUGAAUCAACAUUUCGGUCGGAGUCCACAAAAAGCGGAUGCGCCUCGUGAGUCAUGACCGAUUGCCAAA-UUGAGGCA----- .(((.....)))((((((((.....(((((((....((((......))))((((((((.((((.(.....).))))...)).(....)....))))))))))))))-))))))).----- ( -42.30) >DroAna_CAF1 208314 103 - 1 AGGCAU-AAGCCGCCUCAAUGACUUUUGGCAACAA-AAUGAAUCAACAUUUUGGUUAAAGCUAAAAAACA----------CACUUACGAUAUGACUAAUUGCCGAGAUCCAGCCA----- .(((..-..))).......((..((((((((((((-((((......)))))))(((..........))).----------..................)))))))))..))....----- ( -20.00) >consensus AGGCAUUAAGCCGCCUCAAUGACUUUUGGCAACAA_AAUGAAUCAACAUUUCGGUCAGAGCCAACAAAAAGCGG_CCUGCCUCAUGAGUCAUGACCGAUUGCCAAA_UUGAGGCA_____ .(((.....)))(((((((.....((((((((....((((......))))(((((((.............(((....)))...........))))))))))))))).)))))))...... (-28.57 = -28.75 + 0.18)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:27:26 2006