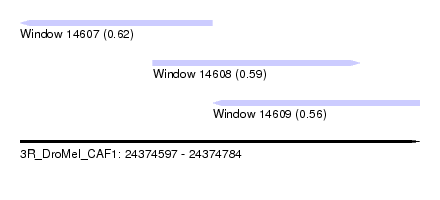
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,374,597 – 24,374,784 |
| Length | 187 |
| Max. P | 0.623291 |

| Location | 24,374,597 – 24,374,687 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 85.48 |
| Mean single sequence MFE | -15.69 |
| Consensus MFE | -12.45 |
| Energy contribution | -12.82 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24374597 90 - 27905053 GCCCCCUCCCCACAGGAUUUC------------UUCGAUGUGUUU-UUUCUC-CCUUUUUUUUGCAUCUCGCGGAUGCUUGCAUCUCGAAAUCAAUCAACUCAA ...............((((((------------...((((((...-...)..-..........(((((.....)))))..)))))..))))))........... ( -14.70) >DroSec_CAF1 196804 101 - 1 GCCCCCUCCCCACAGGAUUUCUUCCCCCACCUCUUCGAUGUGUUU-UUUCUC-CCUUU-UUUUGCAUCUCGCGGAUGCUUGCAUCUCGAAAUCAAUCAACUCAA ..............(((.....)))........(((((.((((..-......-.....-....(((((.....)))))..)))).))))).............. ( -14.71) >DroSim_CAF1 205408 101 - 1 GCCCCCUCCCCACAGGAUUUCUUCCCCCACCUCUUCGAUGUGUUU-UUUCUC-CCUUU-UUUUGCAUCUCGCGGAUGCUUGCAUCUCGAAAUCAAUCAACUCAA ..............(((.....)))........(((((.((((..-......-.....-....(((((.....)))))..)))).))))).............. ( -14.71) >DroEre_CAF1 210829 100 - 1 ACCCCCUCCCCACAGGAUUUCUUCC-CCGCCUCUUCGACGUGUUG-UUUCUC-CCUUU-UUUUGCAUCUCGCGGAUGCUUGCAUCUCGAAAUCAAUCAACUCAA ..............(((.....)))-.......(((((.((((..-......-.....-....(((((.....)))))..)))).))))).............. ( -14.71) >DroYak_CAF1 204229 103 - 1 GCCCCCUCCACACAGGAUUUCUUCCCCCACCUCUGCGAUGUGUUUUUUUCUC-CCUUUUUUUUGCAUCUCGCGGAUGCUUGCAUCUCGAAAUCAAUCAACUCAA ...............((((((.............((((.((((.........-..........)))).))))(((((....))))).))))))........... ( -18.51) >DroAna_CAF1 199879 95 - 1 ACCCCAACCCCCGAUGAUUUCUCUGUCCC-------GCUUUGCUU-CUUCGUUUAUUU-UUUUGCAUCUCGCGGAUGCUUGCAUCUCGAAAUCAAUCAACUCAA ............(((((((((.......(-------((..(((..-............-....)))....)))((((....))))..)))))).)))....... ( -16.77) >consensus GCCCCCUCCCCACAGGAUUUCUUCCCCCACCUCUUCGAUGUGUUU_UUUCUC_CCUUU_UUUUGCAUCUCGCGGAUGCUUGCAUCUCGAAAUCAAUCAACUCAA ...............((((((..............(((.((((....................)))).))).(((((....))))).))))))........... (-12.45 = -12.82 + 0.36)



| Location | 24,374,659 – 24,374,756 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -25.72 |
| Energy contribution | -25.96 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24374659 97 + 27905053 CAUCGAA------------GAAAUCCUGUGGGGAGGGGGCAA-AGAUUCCACUCGGGUGGUCCUCCUAUUGGAAGAUUCACGUUGUAUACCCUAUAGUGAUGAUAUUAUA .......------------...(((((((((((.(((((...-.....)).)))..((((((.(((....))).)).))))........)))))))).)))......... ( -26.40) >DroSec_CAF1 196865 110 + 1 CAUCGAAGAGGUGGGGGAAGAAAUCCUGUGGGGAGGGGGCAAGAUAUUCCACUCGGGUGGUCUUCCUUCUGGAAGAUUCACGUUGUAUACCCUAUAGUAAUGGUGUUAUA ((((.....))))............((((((((.(((((.((....)))).)))..((((((((((....)))))).))))........))))))))............. ( -30.90) >DroSim_CAF1 205469 110 + 1 CAUCGAAGAGGUGGGGGAAGAAAUCCUGUGGGGAGGGGGCAAGAGAUUCCACUCCGGUGGUCUUCCUUCUGGAAGAUUCACGUUGUAUACCCUAUAGUAAUGGUAUUAUA ((((.....))))............((((((((.((((.............)))).((((((((((....)))))).))))........))))))))............. ( -32.02) >DroEre_CAF1 210890 109 + 1 CGUCGAAGAGGCGG-GGAAGAAAUCCUGUGGGGAGGGGGUGAAAUAUUCCACUCGGGUGGUCUUUCUUCUGGAAGAUCCACGCUGCAUACCCUAUAGCGAUGGCGUUCCA ((((.....))))(-(((.(..(((((((((((...(((((........)))))(.((((((((((....)))))).)))).)......)))))))).)))..).)))). ( -44.80) >DroYak_CAF1 204292 103 + 1 CAUCGCAGAGGUGGGGGAAGAAAUCCUGUGUGGAGGGGGCGAAAUAUUCCACUCGGGUGGUCUUCCUUCAAGAAGAUCCGCGCUGUAUACCCUAUAGCGAUGG------- ((((((.((..((((((((((...((((.((((((...........)))))).))))...))))))))))......)).))((((((.....)))))))))).------- ( -42.50) >consensus CAUCGAAGAGGUGGGGGAAGAAAUCCUGUGGGGAGGGGGCAAAAUAUUCCACUCGGGUGGUCUUCCUUCUGGAAGAUUCACGUUGUAUACCCUAUAGUGAUGGUAUUAUA ((((.....))))............((((((((..(.(((........((....))((((((((((....)))))).))))))).)...))))))))............. (-25.72 = -25.96 + 0.24)



| Location | 24,374,687 – 24,374,784 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.09 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -13.76 |
| Energy contribution | -13.45 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24374687 97 - 27905053 ----------GAUAU--CAUGAUUUUGUAAUCCAAGAAUAUAUAAUAUCAUCACUAUAGGGUAUACAACGUGAAUCUUCCAAUAGGAGGACCACCCGAGUGGAAUCU-UU ----------(((((--.((((((((........))))).))).))))).(((((...((((.(((...)))..((((((....))))))..)))).))))).....-.. ( -22.30) >DroSec_CAF1 196905 108 - 1 UAAUAUCAUUGAUAU--CCUGAUUUUCUAAUCGAAGAAUAUAUAACACCAUUACUAUAGGGUAUACAACGUGAAUCUUCCAGAAGGAAGACCACCCGAGUGGAAUAUCUU .....((((((((((--((((.(((((.....))))).....(((.....)))...)))))))).))..)))).((((((....))))))((((....))))........ ( -21.60) >DroSim_CAF1 205509 110 - 1 UUAUAUCCUGGAUAUUUCUGGAUUUUCUAAUCGAAGAAUAUAUAAUACCAUUACUAUAGGGUAUACAACGUGAAUCUUCCAGAAGGAAGACCACCGGAGUGGAAUCUCUU .(((((((((.(((((..(.((((....)))).)..)))))...............)))))))))....(.((.((((((....))))))((((....))))..)).).. ( -25.29) >DroEre_CAF1 210929 95 - 1 AGAUAUGAUAG--------------GAUACUA-CAUACUAUGGAACGCCAUCGCUAUAGGGUAUGCAGCGUGGAUCUUCCAGAAGAAAGACCACCCGAGUGGAAUAUUUC ...((((.(((--------------....)))-))))((.(((((..(((.((((.((.....)).)))))))...)))))..)).....((((....))))........ ( -22.50) >consensus U_AUAUCAU_GAUAU__C_UGAUUUUCUAAUCGAAGAAUAUAUAACACCAUCACUAUAGGGUAUACAACGUGAAUCUUCCAGAAGGAAGACCACCCGAGUGGAAUAUCUU ..................................................(((((...((((.(((...)))..((((((....))))))..)))).)))))........ (-13.76 = -13.45 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:27:15 2006